Dear Editor:
Papillon-Lefèvre syndrome (PLS) is a rare genetic disorder of skin that runs in families in an autosomal recessive fashion. The typical disease symptoms include the thickening of the skin at foot soles and palms of the hands; along with early and destructive loss of primary as well as permanent teeth
123, The disease incidence rate is 1~4 affected cases in 1 million people while carrier ratio is higher; 2~4 in 1,000 people with no gender discrimination
34. Homozygous mutations in cathepsin C (
CTSC) gene is the only reported cause of PLS. The
CTSC is a small gene mapped to the chromosomal region 11q14.2
56 consisting of 7 exons and 6 introns with a total gene size of 46 kb. It encodes cathepsin C protein also known as dipeptidyl-peptidase 1, a lysosomal cysteine protease. Role of
CTSC mutations in PLS is well established. CTSC mutations are also known to cause phenotypically overlapping disease; Haim-Munk syndrome (HMS) and aggressive periodontitis (API). Around 75 mutations of
CTSC gene have been reported so far. Out of these 75 mutations, most of the mutations (around 97%) were reported to cause PLS phenotype while a few (around 3%) mutations have been associated with families having HMS or API phenotypes.
In this study, we aimed to discover the underlying genetic causes in two large Saudi families showing PLS symptoms (
Fig. 1A, B), using microarray and direct Sanger sequencing. Based on the literature review, to the best of our knowledge, this is the ever first report from Saudi Arabia reporting a novel pathogenic mutation along with a recurrent mutation in
CTSC gene in a PLS affected Saudi family.
The said work was conducted after securing permission from the Ethical Review Committee of College of Medicine, Taibah University (IRB no. 051-02-2017). Pedigrees were drawn after getting the informed consents, to commence the study. We received the patient's consent form about publishing all photographic materials. Clinical evaluation of the family was performed by a dermatologist at Madinah Maternity and Children Hospital Almadinah Almunawwarah.
Five blood samples were taken from individuals (III:1, III:2, IV:1, IV:2, IV:3) of family A while a total of thirteen blood samples were collected from individuals of family B including five affected (IV:3, IV:4, V:2, V:3, V:6) and eight unaffected (III:3, III:4, III:5, III:6, IV:1, IV:2, IV:9, IV:10) individuals, to extract the genomic DNA using QIAquick DNA extraction kit. Quality and integrity of the extracted DNA was checked by Nanodrop spectrophotometer and gel electrophoresis.
DNA of one affected (IV:1) and four unaffected members including both parents (III:1, III:2) and two unaffected siblings (IV:2, IV:3) were used for whole genome single nucleotide polymorphism (SNP) genotyping and Sanger validation following same protocols as used elsewhere
78.
Patients were clinically evaluated by consultant dermatologist, manifesting typical features of PLS, however, interand intrafamilial variability was observed in terms of the degree of involvement of oral and skin conditions. Inflammation and recession of the gums, loosening of the teeth and palmoplantar keratoderma were the main clinical manifestations. Affected individual (IV:1) of family A showed palmoplantar keratosis and severe periodontitis affecting deciduous and permanent dentitions. Affected individual (V:2) of family B showed exfoliated primary teeth, plaque accumulation, hyperkeratosis of the palms, fingers, soles, knees, elbows, and back (
Fig. 1C~F). Hyperkeratotic areas were observed with crustations and fissuring palmar surface of her hands and on plantar surface of her feet.
Homozygosity analysis of SNP genotypes, obtained from 250K Affymetrix array, revealed a homozygous region of approximately 20 Mbs (85 Mb to 105 Mb) on chromosome 11a14.1-q22.3. Unaffected members of the family didn't show any homozygosity in this region.
Homozygous region, identified using genotyping data, contain
CTSC gene.
CTSC is a known candidate gene for PLS, therefore,
CTSC gene was sequenced directly. Bi-directional Sanger sequencing of entire coding regions and splice site sequences of
CTSC gene revealed a novel (c.890G>A) and a recurrent (c.902G>T) missense mutations within exon 7 in families A and B, respectively. The novel mutation g.88294508C>A, at cDNA position 890 is predicted to cause amino acid shift from glycine to aspartate at position 297. The recurrent mutation, identified in patients from family B, in exon 7 of the
CTSC gene was at cDNA position 902, is predicted to change glycine to valine at amino acid position 301 (
Fig. 2).
Various software including SIFT, PolyPhen, MutationTaster, MutationAssesor, PROVEAN, VEST3, and others predicted that both missense variants are in the active site of the CTSC enzyme, and, therefore, are of high functional impact. Prediction software categorized these variants as deleterious, disease causing or damaging (
Table 1). Protein structure stability prediction for mutation using MUpro (
https://www.ics.uci.edu/~baldig/mutation.html) and IMutant2.0 (
http://folding.biofold.org/cgi-bin/i-mutant2.0.cgi) predicted that the variants decreases the stability of protein structure.
PLS, API, and HMS are allelic syndromes. A salient features common in all these phenotypes is early onset of severe periodontitis leading to ultimate tooth loss. The families under investigation in this study segregates PLS and not HMS as hallmarks of HMS, acroosteolysis and arachnodactyly, were absent in all affected members of both families. Mutations in CTSC gene lead to PLS phenotype.
Around 75 mutations including deletion, insertion, frameshift, nonsense, missense, and splice-site have been reported in
CTSC gene so far
69. Though missense mutations are reported to be found in all the coding regions of
CTSC gene, the bulk of these mutations are found in exon 5 to 7 that is responsible for encoding the active part of the CTSC protein involved in active enzyme activity
610. Both variants (c.890G>A and c.902G>T), identified in this study, are present in exon 7 of the
CTSC gene. They are very rare in population and not found in the ESP6500, ExAC and 1,000 genomes databases. Moreover, both mutations change the amino acids (p.G297D and p.G301V) in the functionally active site (peptidase C1A, papain C-terminal) of the CTSC enzyme.
Genetic analysis of sporadic cases and families segregating PLS is helpful in improving the quality of life by early diagnosis and proper treatment of PLS symptoms.
Figures and Tables
Fig. 1
Pedigree charts of two families (A and B) segregating Papillon-Lefevre Syndrome (PLS) and clinical presentation of PLS features in an affected individual from family B. Intraoral appearance showing serious periodontitis, loss of permanent teeth from both jaws, inflammation, and enlargement of the gingiva (C). Hyperkeratotic lesions on the dorsal surface of hands (D) Keratotic, confluent plaques affecting the skin of sole and extending on to the dorsal surface, psoriasis form lesions on the ankle (E and F). +/+, +/−, and −/− shows wild type, carrier and homozygous mutants, repsctively.
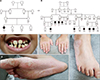
Fig. 2
Partial DNA sequence of the exon 7 of the CTSC gene. (A~C) Shows partial sequence of the affected, carrier and normal individuals of family A. (D~F) Shows partial sequence of the affected, carrier and normal individuals of family B. Arrows indicate position of the mutation.

Table 1
Pathogenicity prediction of variants identified in the cathepsin C gene with various prediction effect softwares







 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download