This article has been
cited by other articles in ScienceCentral.
Dear Editor:
Inherited ichthyoses comprise a large group of cornification maladies characterized by dry, rough and scaly appearance of most or all the integument. Ichthyoses have a heterogenous genetic background with >50 associated genes segregating in autosomal recessive/dominant and Xlinked inheritance pattern. Its subtypes can be distinguished based on the pattern of hyperkeratosis/scaling, mode of inheritance, onset and alterations over time, manifestations in other organs, and family history. Yet, the phenotypic heterogeneity occasionally necessitates laboratory investigations and genetic analysis for proper diagnosis
1. These are categorized as rare disorders having an incidence rate of 5 to 10/100,000 in United States (
http://www.firstskinfoundation.org); however, the prevalence for individual subtypes of ichthyosis vary widely depending on the mode of inheritance and demographic location. Although, data on prevalence of ichthyoses in Pakistani population is not available, a study conducted to analyze harlequin ichthyosis enrolled 45 patients worldwide, 19 (~42%) of whom belonged to Pakistan and its surroundings
2. In this demographic location, the incidence rate of inherited disorders is higher compared to rest of the world due to frequent consanguinity practices, and lack of awareness and genetic counselling
3. Here we report an inbred Pakistani family affected with two different ichthyoses: lamellar ichthyosis (LI) and X-linked ichthyosis (XLI).
The consanguineous family had three patients (V-1, V-2, and V-3) born to phenotypically normal parents (III-1 and IV-1) (
Fig. 1A). Patient V-3 had died during infancy. The appearance and severity of the phenotype in V-1 and V-2 dramatically varied from each other. V-1 had a very severe phenotype of classic LI. Born as collodion baby, V-1 presented generalized, dark-brown, large, plate like adherent scales. Ectropion, eclabium, hypotrichosis, scarring alopecia, palmoplantar hyperkeratosis, and onychauxis were seen. Flexural contractures were observed on axilla, groin, neck, and antecubital fossae (
Fig. 1B~E). The phenotype of V-2 was milder than V-1. V-2 presented generalized, light to dark-brown scales, with mild involvement of face and scalp and normal scalp hair. Other features including nail deformities, ectropion, eclabium, and palmoplantar hyperkeratosis were not observed (
Fig. 1F~H). His phenotype, although different from V-1, was initially categorized as autosomal recessive congenital ichthyosis (of which LI is a subtype) due to the frequently observed phenotypic heterogeneity within patients of a family
14. The study was proceeded after obtaining approval from the institutional review board of the Quaid-i-Azam University (IRB no. 216) and written informed consents about investigating and publishing the data along with photographic material from the enrolled individuals.
Owing to the higher proportion of
TGM1 (which encodes for transglutaminase-1 protein) mutations underlying LI
4,
TGM1 was sequenced in the proband V-1 as done previously
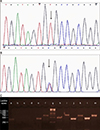
5. Sequencing identified a homozygous missense mutation c.424C>T (p.R142C) in the gene (
Fig. 2A). The p.R142C mutation has often been observed in individuals affected with classic LI (similar to V-1 phenotype) and is found to completely eliminate the enzyme activity in murine epidermis
6. Segregation analysis of the mutation revealed an interesting scenario; the other patient V-2 and the normal individuals III-1, IV-1, and V-4 were heterozygous for the mutant allele (
Fig. 2B). Thus, complete
TGM1 was screened in V-2 to find another variation that could render the protein nonfunctional. However, c.424C>T was the sole divergence from reference sequence of
TGM1 in V-2. The possibility of a dominant negative effect of the mutant protein was eliminated by the normal heterozygous parents. Thus,
TGM1 sequencing remained inconclusive to rule out the single cause of ichthyosis in the family necessitating re-examination of V-2.
Predominance of male patients and sparing of face and palmoplantar surfaces in V-2 clued XLI presentation. About 90% of reported XLI cases occur due to complete or partial deletions in steroid sulfatase gene (
STS)
7, thus,
STS exon-1 and 10 were tested through polymerase chain reaction amplification in V-2 and a normal control individual. Exon-1 was amplified in both samples but exon-10 was not amplified in V-2 indicating a deletion at the 3′ end of
STS (
Fig. 2C). Further screening of the upstream exons and downstream markers RH36507, DXS7434E, and D12S1026, delimited the deletion starting in intron-7 and ending upstream of DXS7434E spanning ~47 kb genomic region. V-1 did not carry the deletion. The identified deletion predicts a stop lost as well as the loss of 218 amino acids at the C-terminal of protein. This mutant protein would lack several important amino acids critical for substrate recognition, transmembrane domain, and active site
8 thus affecting the normal function of the protein.
In the studied family, the phenotypic heterogeneity often observed in autosomal recessive congenital ichthyosis
4 and the two disease entities being related in terms of appearance of the integument were the leading causes of misdiagnosis in the patient V-2. This seemingly uncommon scenario has also been found elsewhere where a family was segregating the most common ichthyoses viz the semi-dominant ichthyosis vulgaris (prevalence 1:250), and XLI (prevalence 1:2,000), either disorder was inherited from a parent
9. Besides, a recent report shows concurrent congenital myopathies and hypophosphatemic rickets in a consanguineous family which was resolved only after being analyzed molecularly through whole exome sequencing
10. With the higher accessibility and advancements in molecular technologies, this scenario is expected to be observed more often especially in areas where consanguinity and marriages within tribe/caste are frequently practiced. The degree of consanguinity and the frequency of the pathogenic allele are directly related to the chances of co-occurrence of inherited diseases
3. Therefore, in addition to phenotypic heterogeneity, the higher chances of co-occurrence of genetic disorders in inbred populations should never be dismissed for accurate diagnosis, as otherwise, the co-occurring features of etiologically different disease entities within the same individual or concurrence of related inherited diseases in members of the same family may lead to misleading diagnosis and genetic counseling. Moreover, a multigene panel covering all the genes related to the disease symptoms is considered the test of choice to avoid the confusion arising from overlapping features of the disorders
410.
To conclude, we identified two Pakistani siblings affected with different types of ichthyoses. The scenario reflects an adverse outcome of the higher consanguinity and sociocultural norms practiced in this demographic location.
Figures and Tables
Fig. 1
(A) Pedigree of the studied family. Squares and circles represent males and females, respectively. Clear and filled symbols are indicative of unaffected and affected status of individuals, respectively. Symbols with crossed lines indicate deceased individuals. Double lines denote consanguineous marriage and an arrow at the left bottom corner of symbol denotes proband. (B~E) Phenotypic appearance at different anatomic sites of the affected individual V-1. (F~H) Phenotypic appearance at different anatomic sites of the affected individual V-2.

Fig. 2
Screening of TGM1 and STS in the affected individuals. (A) Chromatogram demonstrating mutation c.424C>T in exon 3 of TGM1 in V-1 and (B) in V-2, (C) polymerase chain reaction amplification of STS exons using gDNA of affected individuals V-1 and V-2 and an ethnically matched positive control. a=exon 7_V-2 (443 bp), b=exon 8_V-2 (682 bp), c=exon 9_V-2 (485 bp), d=exon 10_V-2 (757 bp), e=exon 7_V-1 (443 bp), f=exon 8_V-1 (682 bp), g=ladder, h=exon 9_V-1 (485 bp), I=exon 10_V-1 (757 bp), j=exon 7_control (443 bp), k=exon 8_control (682 bp), l=exon 9_control (485 bp), m=exon 10_control (757 bp).
ACKNOWLEDGMENT
NK was supported by the Higher Education Commission of Pakistan through indigenous PhD fellowship program, phase II, Batch II.
References
1. Vahlquist A, Fischer J, Törmä H. Inherited nonsyndromic ichthyoses: an update on pathophysiology, diagnosis and treatment. Am J Clin Dermatol. 2018; 19:51–66.

2. Rajpopat S, Moss C, Mellerio J, Vahlquist A, Gånemo A, Hellstrom-Pigg M, et al. Harlequin ichthyosis: a review of clinical and molecular findings in 45 cases. Arch Dermatol. 2011; 147:681–686.
3. Bittles AH, Black ML. Evolution in health and medicine Sackler colloquium: consanguinity, human evolution, and complex diseases. Proc Natl Acad Sci U S A. 2010; 107 Suppl 1:1779–1786.
4. Richard G. Autosomal recessive congenital ichthyosis. In : Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, editors. GeneReviews®. Seattle: University of Washington;2001.
5. Rasheed M, Karim N, Shah FA, Naeem M. Novel TGM1 mutation in a Pakistani family affected with severe lamellar ichthyosis. Pediatr Neonatol. 2018; 59:628–629.

6. Nakagawa N, Yamamoto M, Imai Y, Sakaguchi Y, Takizawa T, Ohta N, et al. Knocking-in the R142C mutation in transglutaminase 1 disrupts the stratum corneum barrier and postnatal survival of mice. J Dermatol Sci. 2012; 65:196–206.

7. Crane JS, Wu B, Paller AS. Ichthyosis X-Linked. Treasure Island: StatPearls Publishing;2019.
8. Ghosh D. Three-dimensional structures of sulfatases. Methods Enzymol. 2005; 400:273–293.

9. Rice RH, Bradshaw KM, Durbin-Johnson BP, Rocke DM, Eigenheer RA, Phinney BS, et al. Distinguishing ichthyoses by protein profiling. PLoS One. 2013; 8:e75355.

10. Lal D, Neubauer BA, Toliat MR, Altmüller J, Thiele H, Nürnberg P, et al. Increased probability of co-occurrence of two rare diseases in consanguineous families and resolution of a complex phenotype by next generation sequencing. PLoS One. 2016; 11:e0146040.







 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download