Abstract
Neddylation is a post-translational protein modification process. MLN4924 is a newly discovered pharmaceutical neddylation inhibitor that suppresses cancer growth with several cancer types. In our study, we first investigated the effect of MLN4924 on colon cancer cells (HCT116 and HT29). MLN4924 significantly inhibited the neddylation of cullin-1 and colon cancer cell growth in a time and dose-dependent manner. MLN4924 induced G2/M cell cycle arrest and apoptosis in HCT116 and HT29 cells. Moreover, MLN4924 also triggered autophagy in HCT116 and HT29 cells via suppressing the PI3K/AKT/mTOR pathway. Inhibiting autophagy by autophagy inhibitor 3-MA or ATG5 knockdown reversed the function of MLN4924 in suppressing colon cancer cell growth and cell death. Interestingly, MLN4924 suppresses colon cell growth in a xenograft model. Together, our finding revealed that blocking neddylation is an attractive colon cancer therapy strategy, and autophagy might act as a novel anti-cancer mechanism for the treatment of colon cancer by MLN4924.
Colon cancer involves aberrant growths in the appendix, colon and rectum and is the third most commonly seen malignancy and one of the most common causes of cancer-related death in worldwide [1]. Despite the remarkable development in prevention, diagnostic techniques and improvements in chemotherapy, the median overall survival period of colon cancer patients with metastatic is only 24 months [23]. Therefore, it is urgent to develop new therapeutic agents and anticancer targets to improve the treatment of colon cancer. Accumulating evidence demonstrated that autophagy is an important cellular process in cancer cell survival and cell death [4]. Autophagy is a conserved homeostatic mechanism of lysosomal degradation that characterized by formation of double or multi-membrane vesicles in cytosol (autophagosomes). These autophagosomes, encapsulating bulk cytoplasm and organelle, mature by fusing with the endocytic compartments and then fusing with lysosomal compartment to form autolysosomes, in which the cargo is degraded by acidic lysosomal hydrolases [5]. It has been well documented that autophagy is involved in colon cancer cells proliferation, migration, invasion and chemotherapy sensitivity [6].
Neddylation adds the ubiquitin-like molecule NEDD8 to substrates and thus regulates their conformation, localization, stability and function [7]. Cullin-family proteins are well-known substrates of neddylation that are scaffolds of multi-unit cullin-RING E3 ligase (CRL) complexes [89]. NEDD8 conjugation to cullins induces conformational changes and activation of CRL to ubiquitin/proteasome-dependently degrade a number of proteins that are crucial for inhibiting cell proliferation and survival (such as p21 and p27) [810]. Therefore, the inactivation of CRL, through inhibiting neddylation pathway, has been a therapy strategy for suppressing the growth of cancer cells [1112].
MLN4924 is a newly discovered pharmaceutical neddylation inhibitor and developed as an anti-cancer drug [13]. MLN4924 efficiently blocks neddylation of cullins, resulting in inactivation of CRLs [8]. Accumulation of CRLs substrates induces DNA damage response, cell cycle arrest, apoptosis and senescence in several cancer types, including liver cancer, gastric cancer, osteosarcoma and colon cancer [101415]. Recently, accumulating evidence suggests that autophagy may be involved in the induction of cancer cell proliferation and survival upon cellular stress [16]. However, it remains unclear whether autophagy responds upon inhibition of neddylation and regulates cell growth in colon cancer. In the present study, it's showed that MLN4924 significantly inhibited neddylation of cullin-1 and cell growth in colon cancer. MLN4924 effectively induced G2/M cell cycle arrest and apoptosis in a time-dependent manner. Moreover, MLN4924 also triggers autophagy via inhibiting PI3K/Akt/mTOR pathway. The inhibition of autophagy reversed MLN4924-induced cell growth in colon cancer cells. Our findings provide evidence that MLN4924 might be used as a novel class anti-cancer agent in the treatment of colon cancer.
Colon cancer cell lines, HCT116 and HT29, were purchased from American Type Culture Collection (ATCC, USA). Cells were cultured in Dulbecco's Modified Eagle's medium (GIBCO, USA) containing 10% fetal bovine serum (FBS, Hyclone, USA) and 1% penicillin-streptomycin solution (Beyotime, China), in a humiliated incubator with 5% CO2 at 37℃.
MLN4924 (Sigma, USA) was dissolved in DMSO to make a stock solution and further diluted to a corresponding concentration with complete culture medium. The final DMSO concentration was less than 0.01% (v/v). HCT116 and HT29 cells were treated with 0.3 µM MLN4924 for different time periods (12, 24, 48 h) or different concentration of MLN4924 (0, 0.01, 0.3, 1, 3 and 10 µM) for 24, 48 or 72 h. For inhibition of autophagy, cells were treated with 3-MA or co-treated with MLN4924 (0.3 µM) for 24, 48 or 72 h. Then the cells were used for corresponding experiments.
BrdU cell proliferation ELISA kit (Abcam, USA) was used for BrdU assay. Briefly, HCT116 and HT29 cells were seeded in 96-well culture plates for 24 h. After different treatments, cells were incubated with 10 µM BrdU for 2 h at 37℃. When the incubation period ended, the cells were fixed and denatured DNA by fixing solution, then incubated with anti-BrdU antibody for 1 h at room temperature (RT). After washed with PBS, cells were incubated with HRP-conjugated secondary antibody for 1 h at room temperature. Subsequently, cells washed with PBS and incubated with TMB solution. Finally, after color developed and then added the stop solution and measured the OD value in each well at 450 nm.
To determine colony formation, HCT116 and HT29 cells were cultured in complete medium and seeded in a 6-well culture plate (containing 1,000 cells) and cultured with MLN4924 (0.3 µM) for 14 days. Then, cells were stained with 0.1% crystal violet and counted by light microscopy. Three independent experiments were performed.
For cell cycle assays, after being treated with MLN4924 (0.3 µM) for 48 h, HCT116 and HT29 cells were harvested and fixed in 70% ethanol overnight at 4℃ and stained with PI solution (50 µg/ml, sigma) containing RNase (10 µg/ml, sigma) for 30 min at room temperature. Then cell cycle was determined by BD FACSAria™ flow cytometry (Becton Dickinson, San Jose, CA, USA).
For apoptosis assays, after being treated with MLN4924 (0.3 µM) for 12, 24 or 48 h, HCT116 and HT29 cells were harvested with 0.25% trypsin without EDTA, washed twice with cold PBS and re-suspended in 500 µl binding buffer. Then cells were incubated with 5 µl annexinV-FITC (40 µg/ml, Becton Dickinson) and 5 µl propidium iodide (40 µg/ml) in the dark for 10 min at room temperature and detected by the flow cytometry.
HCT116 and HT29 cells were transfected with mRFP-GFP-tagged LC3 vector (a gift from Yoshimori [17]) using lipo2000 (Invitrogen, USA). Then, after different treatments, a confocal microscope (Olympus, Japan) was used for the detection of autophagy. Quantification of autophagic vacuoles was analyzed by calculating the number of LC3 puncta from three fields containing more than 5 randomly selected microscopy-captured images, each comprising between 2 and 8 cells. Autophagosomes were detected as RFP+GFP+ (yellow dot), while mature, autolysosomal organelles were detected as RFP+GFP (red-only dot) [1819].
For ultra-structural analysis, transmission electron microcopy (TEM) was performed. After different treatments, HCT116 and HT29 cells were fixed and embedded. Thin sections (90 nm) were cut and analyzed at 80 kV with a JEOL 1200EX transmission electron microscope. Autophagosomes were defined as double membrane vacuoles measuring 0.1 or 2.0 µm.
The following lentiviral constructs were purchased from Santa Cruz: ATG5 shRNA (sc-41445-V) and noncoding shRNA (sc-108080). Lentiviral particles were used to directly infect HCT116 and HT29 cells, and then stable clones were selected using puromycin (Sigma). The selected cell populations were subjected to western blot to determine the silencing efficiency.
Western blotting was conducted as previously described with some modifications [20]. After different treatments, cells were harvested using scraper and lysed in RIPA (Beyotime, China). Cell lysates were centrifuged at 14,000×g for 15 min at 4℃ and supernatants was subjected to western blotting analysis. Proteins (20 to 100 µg) were loaded on 8–15% SDS-PAGE and transferred to nitrocellulose membrane (Applygen Technologies Inc. Beijing, China). The membranes were blocked with 5% non-fat milk in TBST for 2 h and incubated with primary antibodies (anti-p-H2A, anti-T-H2A, anti-p-CHK2, anti-T-CHK2, anti-p21, anti-p27, anti-caspase-3, anti-PARP, anti-LC-3, anti-cullin-1, anti-ATG5, anti-p62, anti-p-70S6K, anti-p-mTOR, anti-p-AKT and anti-GAPDH antibodies, Santa cruz, USA). After being washed three times with TBST, the membranes were incubated for 1 h with corresponding horseradish peroxidase-conjugated secondary antibodies (Santa cruz, USA). The blots were detected using ECL Western Blot Substrate kit (Biotech, USA). The load protein was normalized to GAPDH. The blots were quantified by ImageJ software (National Institute of Mental Health, Bethesda, MD, USA).
Mice were randomly divided into two groups (ten mice per group): (a) vehicle treatment, (b) intraperitoneal administration with MLN4924 (60 mg/kg) three times a week. At five-day intervals, mice were examined for tumor growth or survival. Tumor diameter was measured with a caliper, and tumor volume was calculated based on the following formula: volume=(greatest diameter)×(smallest diameter)2/2. Six weeks after treatment, mice were sacrificed, and tumor sections (5 µm) were subject to either hematoxylin-eosin (H&E) staining or terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay. TUNEL-positive (brown staining) cells were characterized as apoptotic cells, and 10 randomly selected microscopic fields in each group were examined to calculate the ratio of TUNEL-positive cells. Surviving mice were sacrificed under anesthesia. The animal experiment was conducted at 210 Hospital of Chinese People's Liberation Army (Dalian, China), complying with the national guidelines for the care and use of laboratory animals. All animal experiments were approved by the experimental animal ethics committee of 210 Hospital of Chinese People's Liberation Army.
Data were presented as means±SD of at least three independent experiments and statistically analyzed by one-way analysis of variance (ANOVA) followed by Tukey's multiple comparison test or an independent sample t-test using the SPSS Statistics 19.0 software. The level of significance was based on the probability of p<0.05, p<0.01 and p<0.001.
As Nedd8-Activating Enzyme Inhibitor, MLN4924 could effectively inhibit neddylation of cullins [13]. Therefore, we first confirmed the effect of MLN4924 on cullins-1 neddylation using western blot analysis. As shown in Fig. 1A, 0.3 µM MLN4924 completely blocked neddylation of cullins-1 in HCT116 and HT29 cells. We next determined the ability of MLN4924 to inhibit cell proliferation by treating HCT116 and HT29 with different concentrations of the drug (0.01, 0.1, 0.3, 1, 3, 10 µM ) for 24 to 72 h. The proliferation of HCT116 and HT29 was significantly suppressed by MLN4924 in a dose and time-dependent manner (Fig. 1B). Moreover, MLN4924 also notably suppressed the colongenic ability of these cells (Fig. 1C).
It was reported that MLN4924 could enhance radiation or chemotherapy-induced colon cancer cell apoptosis, cell cycle arrest and DNA damage response [2122]. Similarly, our results showed that MLN4924 induced DNA damage response in colon cancer cells, reflecting in appearance phosphorylation of H2A and CHK2. As the substrates of CRL, p21 and p27 expression was significantly up-regulated during the MLN4924 treatment (Fig. 2A). The flow cytometry results showed that MLN4924 effectively triggered G2/M cell cycle arrest and apoptosis in HCT116 and HT29 cells (Figs. 2B and C). Correspondingly, the increase of cleaved caspase-3 and cleaved PARP also confirmed that MLN4924 induced apoptosis in colon cancer cells (Fig. 2D).
Whether MLN4924 triggers autophagy in colon cancer cells remains unclear. We thus performed the ultra-structural analysis by transmission electron microscope. As shown in Fig. 3A, the double-membraned autophagosome was observed in MLN4924 treated cells rather than control cells. Western blot results showed a time-dependent conversion of LC3 I to LC3 II and decrease of p62, two classic measurements of autophagy [23]. To elucidate the role of MLN4924 in promoting the autophagic process, a tandem fluorescent GFP-mRFP-LC3 marker was utilized to analyze autophagosome maturation [18]. Autophagosomes show red and green fluorescence, while autolysosomes show only red fluorescence, due to the fact that GFP lost its fluorescence from deprotonation in the acidic lysosomes. Therefore, autophagosomes display yellow dot (REP+GFP+), and maturation of autophagosome displays a dramatic increase in the number of red fluorescenceonly autolysosomes (RFP+GFP-, red dot). As shown in Fig. 3C, an increase of red-only dot fluorescence was observed in both HCT116 and HT29 cells treated with MLN4924, indicating that MLN4924 effectively enhanced autophagosome maturation in colon cancer cells.
PI3K/Akt/mTOR pathway is a classic modulator of autophagy [24]. Hence, the key proteins in the pathway were investigated. After MLN4924 treatment, the expression of p-AKT, p-mTOR and p-70S6K decreased in a time-dependent manner in HCT116 and HT29. Data demonstrated that MLN4924 induced colon cancer cell autophagy via activating PI3K/Akt/mTOR pathway (Fig. 4).
To further investigate the role of autophagy in colon cancer cell growth and cell death, we first utilized autophagy inhibitor 3-MA to block the autophagy process and then determined the cell proliferation under MLN4924 treatments. BrdU assay results showed that MLN4924 effectively inhibited cell proliferation both in HCT116 and HT29 cells; and co-treated with 3-MA, MLN4924 significantly reduced the inhibition effect (Fig. 5A). Moreover, we evaluated whether the suppression of autophagy, using pharmacological and genetic inhibition, would interfere with MLN4924-induced apoptosis. Figs. 5B and C show that pretreatment with 3-MA significantly decreased MLN4924-induced apoptosis in HCT116 and HT29 cells compared with MLN4924 alone. 3-MA alone did not induce apoptosis in HCT116 and HT29 cells. Fig. 5B demonstrates that 3-MA treatment decreased the accumulation of LC3II and inhibited p62 degradation in HCT116 and HT29 cells treated with MLN4924. In addition, MLN4924-induced Caspase-3 and PARP cleaved in HCT116 and HT29 cells were profoundly attenuated by treatment with 3-MA (Fig. 5B). Similar results were observed in the data of FACS analysis (Fig. 5C). Then, we blocked the autophagy pathway via lentivirus suppress of autophagy gene ATG5 to evaluate cell proliferation. As shown in Fig. 5D, both HCT116 and HT29 cell lines with stable depletion of ATG5 (shATG5➀ and shATG5➁) attenuated the MLN4924-induced inhibition of cell proliferation. In addition, colon cancer cell lines with stable depletion of ATG5 that displayed decreased apoptosis after a 48 h MLN4924 treatment are compared with cells stably infected with lentivirus targeting control shRNA by western blot in Fig. 5E. Similar results were obtained by FACS analysis (Fig. 5F). Efficient knockdown of ATG5 in HCT116 and HT29 cells were attested by western blot (Fig. 5E). In summary, inhibiting autophagy pharmacologically and genetically attenuating MLN4924-induced apoptosis, suggests that autophagy may play an important role in supporting the full development of apoptosis phenotype induced by MLN4924 in colon cancer cells.
We further examined the effect of MLN4924 on colon cell growth in xenograft derived from HCT116 and HT29 cells. MLN4924 markedly abrogated the progression of established tumors as compared with controls (Fig. 6A, p<0.05). Further histologic examination and TUNEL stain of harvested tumors showed increased inflammatory cell infiltration and cell death in tumors treated with MLN4924 (Fig. 6B). No weight loss was observed in control mice injected with MLN4924, indicating that this treatment was well tolerated (data not shown).
Colon cancer is the third most common malignancy and one of the most common causes of cancer-related death in worldwide [1]. Overcoming multidrug resistance is a crucial clinical issue also a critical challenge in colon cancer therapy even after surgical resection. Therefore, it's urgent to develop novel anti-cancer drugs for colon cancers. Recently, blocking protein neddylation pathway has emerged as an attractive anti-cancer approach. As a NAE Inhibitor, MLN4924 shows anti-proliferation, apoptotic and autophagic effects in multiple cancer types via inhibiting the neddylation pathway. In this study, we investigated the anticolon cancer activities of MLN4924 and revealed the underlying mechanism. We showed that MLN4924 significantly inhibited cell growth in colon cancer. MLN4924 also effectively induced cell cycle arrest, apoptosis and autophagy. The trigger of autophagy by MLN4924 is associated with inhibition of PI3K/Akt/mTOR pathway. Moreover, inhibition of autophagy reversed MLN4924-induced cell growth inhibition and cell death in colon cancer cells. Our findings provide evidence that MLN4924 might act as a novel anti-cancer agent in the treatment of colon cancer.
Numerous research findings demonstrated that neddylation is hyper-activated in cancers leading to elevated global neddylation of substrates, including cullins, to promote the ubiquitin-dependent degradation of tumor suppressors and facilitate carcinogenesis and cancer development [12]. Therefore, inhibition of cullins neddylation by the NAE inhibitor MLN4924 leads to accumulation of tumor suppressive CRL substrates, and triggers inhibition of tumor growth and metastasis [1325]. For instance, in liver cancer and gastric cancer, utilization of MLN4924 effectively suppressed cancer cell growth and migration by inducing apoptosis and cell cycle arrest [14]. In colon cancer, MLN4924 sensitized colon cancer cells to radiation by enhancing radiation-induced G2/M arrest and apoptosis and DNA damage response [22]. However, the role of MLN4924 in colon cancer cell death and autophagy is not totally understood. In the present study, cullin-1 neddylation was effectively inhibited by MLN4924 as expected. MLN4924 inhibited cell proliferation and the clonogenic ability in colon cancer cell lines HCT116 and HT29 (Fig. 1). Similarly, our findings also confirmed that MLN4924 triggered cell cycle arrest and apoptosis, accompanied by a series of DNA damage response and cell cycle and apoptosis-associated proteins expression change. Moreover, the TEM and confocal microscopy results showed that MLN4924 acted as an inducer of autophagy in colon cancer cells (Fig. 3).
Autophagy is an intracellular degradative process in which damaged organelles and long-lived proteins are degraded for maintaining cellular homeostasis [26]. The hallmark of autophagy is the formation of double- or multi-membrane autophagosomes that fuses with lysosomes for the degradation of its contents [27]. Autophagy happens in multiple biological processes [428]. Although many signaling pathways are reported to regulate autophagy process, the PI3K/AKT/mTOR pathway is a classic negative regulator of autophagy. Kinase mTOR is a component of two mTOR complexes, mTORC1 and mTORC2. mTORC1 is a major negative regulator of autophagy and PI3K/AKT pathway is an major upstream modulator of mTORC1 [24]. Our study revealed that MLN4294-induced autophagy is mediated by inactivation of PI3K/AKT/mTOR pathway (reflected by decreased levels of p-AKT, p-mTOR and p-70S6K) (Fig. 4).
Besides PI3K/AKT/mTOR pathway, autophagy process is tightly regulated by a series of core autophagy-related (ATG) proteins [29]. During autophagy, LC3I is converted to lapidated LC3II and associated with autophagic membrane. ATG5 is essential for the conversion of LC3I to LC3II and for the location of LC3II to autophagosomes [30]. 3-MA is a frequently-used autophagy inhibitor that suppresses the activity of PI3K and the formation of pre-autophagosome [31]. Therefore, to further explore the role of autophagy in MLN4924-induced colon cancer cell growth inhibition, we blocked autophagy with 3-MA or knockdown ATG5 and then assessed cell proliferation and apoptosis. Interestingly, after blocking autophagy, the inhibition effect of MLN4924 on colon cancer cell growth was reversed, indicating that this inhibition effect was attributed to autophagy. However, the MLN4924-induced autophagy was considered as self-protective mechanism in several cancer cells, including liver cancer and breast cancer [32]. The biological role of autophagy in regulation of cell survival in response to cellular stress is controversial. Some studies indicated that autophagy primarily serves as a pro-survival mechanism against cellular stress while others showed that autophagy could cause autophagic cell death [3334]. Thus, in colon cancer cell, MLN4924-induced autophagy eventually caused autophagic cell death, resulting in cell growth inhibition. In this study we provided evidence that MLN4924 markedly abrogated the progression of established tumors. Our data suggested that MLN4924 may represent a promising therapeutic approach in colon cancer.
Figures and Tables
Fig. 1
MLN4924 inhibits the neddylation of cullins-1 and the cell proliferation in colon cancer cells
(A) MLN4924 inhibits cullins-1 neddylation. HCT116 and HT29 cells were treated with vehicle or 0.3 µM MLN4924 for 12, 24 or 48 h. The expression of cullins-1 and cullins-1-nedd8 was determined by western blot. GAPDH was used for loading control. (B) HCT116 and HT29 cells were treated with vehicle or different concentration of MLN4924 for 24, 48 or 72 h. The cell proliferation was measured by BrdU assays. (C) MLN4924 inhibited cell clonogenic ability. HCT116 and HT29 cells were seeded in 6-well culture plate at 1000 cells per well and treated with vehicle or MLN4924 for 14 days, followed by crystal violet staining and colony counting. The statistical analysis results were shown in the right panels. All values were represented as means±SD calculated from three independent experiments. ***p<0.001.
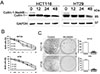
Fig. 2
MLN4924 induced G2 cell cycle arrest and apoptosis in colon cancer cells
HCT116 and HT29 cells were treated with vehicle or 0.3 µM MLN4924 for 12, 24 or 48 h. The expression of p-H2A, T-H2A, p-CHK2, T-CHK2, p21 and p27 was determined by western blot. (B) HCT116 and HT29 cells were treated the same as in (A) and were analyzed by FACS after staining with propidium iodide for cell cycle analysis at 24 h. (C) HCT116 and HT29 cells were treated the same as in (A) and analysis of apoptosis by FACS using AnnexinV/PI double-staining at 12, 24 or 48 h. (D) HCT116 and HT29 cells were treated the same as in (A). The expression of cleaved caspase-3 and cleaved PARP was determined by western blot. All values were represented as means±SD calculated from three independent experiments. **p<0.01, ***p<0.001.

Fig. 3
MLN4924 induced autophagy in colon cancer cells
(A) HCT116 and HT29 cells were treated with vehicle or 0.1 µM MLN4924 for 48 h, then they were analyzed by transmission electron micrograph (TEM), the black arrows indicated the autophagosomes. (B) HCT116 and HT29 cells were treated with vehicle or 0.3 µM MLN4924 for 12, 24 or 48 h. The expression of LC3 and p62 was determined by western blot. GAPDH was used for loading control. (C) HCT116 and HT29 cells were transfected with GFP-mRFP-LC3 vector then they were treated with vehicle or 0.3 µM MLN4924 for 24 h. Images were took with a confocal microscope and the merged region enclosed within the white square has been enlarged in the right panel.
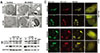
Fig. 4
The PI3K/Akt/mTOR pathway contributes to MLN4924 triggered autophagy
HCT116 and HT29 cells were treated with vehicle or 0.3 µM MLN4924 for 12, 24 or 48 h. The expression of p-AKT, p-mTOR and p-70S6K were determined by western blot. GAPDH was used for loading control.
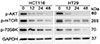
Fig. 5
Autophagy is involved in MLN4924-induced cell growth inhibition in colon cancer cells.
(A) HCT116 and HT29 cells were treated with 3-MA (3 mM) in the absence or presence of 0.3 µM MLN4924 for 24, 48 or 72 h, the cell proliferation was measured by BrdU assays. (B) HCT116 and HT29 cells were treated the same as in (A) for 48 h, The expression of LC3, p62, cleaved caspase-3 and cleaved PARP was determined by western blot. GAPDH was used for loading control (C) HCT116 and HT29 cells were treated the same as in (A) for 48 h, double stained with Annexin V/PI and analyzed by FACS. (D-F) HCT116 and HT29 cell lines with stable depletion of ATG5 were treated with vehicle or 0.3 µM MLN4924 for 48 h. (D) The cell proliferations was measured by BrdU assays. (E) The expression of ATG5, cleaved caspase-3 and cleaved PARP was determined by western blot. GAPDH was used for loading control. (F) Double stained with Annexin V/PI and analyzed by FACS. All values were represented as means ± SD calculated from three independent experiments. ***p<0.001.
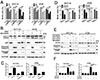
Fig. 6
MLN4924 suppresses colon cancer cell growth in mouse tumor xenograft.
Six weeks old female nude mice bearing HCT116 and HT29 tumors were randomly divided into two groups (ten mice per group): (A) vehicle control, (B) intraperitoneal administration with MLN4924 (60 mg/kg) three times per week. (A) Tumor volumes were measured at 5-day intervals for 40 days and expressed as the mean±SD (n=10) in tumor volume-time curves. (B) six weeks after treatment, tumor tissue samples from each treatments group was subjected to either hematoxylin-eosin (H&E) staining or TUNEL assay.

ACKNOWLEDGEMENTS
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Notes
Author contributions J.G. conceived of the study and designed the assays. Y.L. and B.L. performed major experiments. K.H. and Y.X. performed histological analyses; Y.M. and Z.J. analyzed data and performed the statistical analysis. Y.L. contributed reagents/materials/analysis tools. Y.L. and J.G. wrote and edited the manuscript. All authors read and approved the final manuscript.
References
1. Maxwell PJ IV, Camp ER. Current controversies in the management of colon and rectal cancer. Cham: Springer International Publishing;2016.
2. Petrelli F, Barni S. Correlation of progression-free and post-progression survival with overall survival in advanced colorectal cancer. Ann Oncol. 2013; 24:186–192.

3. Heervä E, Lavonius M, Jaakkola P, Minn H, Ristamäki R. Overall survival and metastasis resections in patients with metastatic colorectal cancer using electronic medical records. J Gastrointest Cancer. 2018; 49:245–251.

4. Kenific CM, Debnath J. Cellular and metabolic functions for autophagy in cancer cells. Trends Cell Biol. 2015; 25:37–45.

5. Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT, Liu B, Bao JK. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012; 45:487–498.

6. Gil J, Pesz KA, Sąsiadek MM. May autophagy be a novel biomarker and antitumor target in colorectal cancer? Biomark Med. 2016; 10:1081–1094.

7. Watson IR, Irwin MS, Ohh M. NEDD8 pathways in cancer, Sine Quibus Non. Cancer Cell. 2011; 19:168–176.

8. Merlet J, Burger J, Gomes JE, Pintard L. Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell Mol Life Sci. 2009; 66:1924–1938.

9. Sui Y, Liu Y, Xu G. A lysine-to-arginine mutation on NEDD8 markedly reduces the activity of cullin RING E3 ligase through the impairment of neddylation cascades. Biochem Biophys Res Commun. 2015; 461:653–658.

10. Xu J, Li L, Yu G, Ying W, Gao Q, Zhang W, Li X, Ding C, Jiang Y, Wei D, Duan S, Lei Q, Li P, Shi T, Qian X, Qin J, Jia L. The neddylation-cullin 2-RBX1 E3 ligase axis targets tumor suppressor RhoB for degradation in liver cancer. Mol Cell Proteomics. 2015; 14:499–509.

11. Wang Y, Luo Z, Pan Y, Wang W, Zhou X, Jeong LS, Chu Y, Liu J, Jia L. Targeting protein neddylation with an NEDD8-activating enzyme inhibitor MLN4924 induced apoptosis or senescence in human lymphoma cells. Cancer Biol Ther. 2015; 16:420–429.

12. Zhao Y, Morgan MA, Sun Y. Targeting Neddylation pathways to inactivate cullin-RING ligases for anticancer therapy. Antioxid Redox Signal. 2014; 21:2383–2400.

13. Nawrocki ST, Griffin P, Kelly KR, Carew JS. MLN4924: a novel firstin-class inhibitor of NEDD8-activating enzyme for cancer therapy. Expert Opin Investig Drugs. 2012; 21:1563–1573.

14. Lan H, Tang Z, Jin H, Sun Y. Neddylation inhibitor MLN4924 suppresses growth and migration of human gastric cancer cells. Sci Rep. 2016; 6:24218.

15. Zhang Y, Shi CC, Zhang HP, Li GQ, Li SS. MLN4924 suppresses neddylation and induces cell cycle arrest, senescence, and apoptosis in human osteosarcoma. Oncotarget. 2016; 7:45263–45274.

16. Ryabaya OO, Egorova AV, Stepanova EV. The role of autophagy in mechanisms of tumor cell death. Biol Bull Rev. 2015; 5:579–588.

17. Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescenttagged LC3. Autophagy. 2007; 3:452–460.

18. Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008; 4:151–175.
19. Jiang K, Liu M, Lin G, Mao B, Cheng W, Liu H, Gal J, Zhu H, Yuan Z, Deng W, Liu Q, Gong P, Bi X, Meng S. Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death. Oncotarget. 2016; 7:25652–25667.

20. Deng S, Tang S, Zhang S, Zhang C, Wang C, Zhou Y, Dai C, Xiao X. Furazolidone induces apoptosis through activating reactive oxygen species-dependent mitochondrial signaling pathway and suppressing PI3K/Akt signaling pathway in HepG2 cells. Food Chem Toxicol. 2015; 75:173–186.

21. Zheng W, Luo Z, Zhang J, Min P, Li W, Xu D, Zhang Z, Xiong P, Liang H, Liu J. Neural precursor cell expressed, developmentally downregulated 8-activating enzyme inhibitor MLN4924 sensitizes colorectal cancer cells to oxaliplatin by inducing DNA damage, G2 cell cycle arrest and apoptosis. Mol Med Rep. 2017; 15:2795–2801.

22. Wan J, Zhu J, Li G, Zhang Z. Radiosensitization of human colorectal cancer cells by MLN4924: an inhibitor of NEDD8-activating enzyme. Technol Cancer Res Treat. 2016; 15:527–534.
23. Koskela A, Reinisalo M, Kaarniranta K. Taking a roller coaster ride with autophagy markers p62 and LC3. Acta Ophthalmol. 2016; 94:S256.

24. Heras-Sandoval D, Pérez-Rojas JM, Hernández-Damián J, Pedraza-Chaverri J. The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cell Signal. 2014; 26:2694–2701.

25. Jia L, Li H, Sun Y. Induction of p21-dependent senescence by a NAE inhibitor, MLN4924 as a mechanism of growth suppression. Neoplasia. 2011; 13:561–569.
26. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008; 451:1069–1075.

27. Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal. 2011; 14:2201–2214.

29. Deegan S. The molecular characterization of ER stress-induced autophagy and cell death [PhD dissertation]. Ireland: NUI Galway;2012.
30. Kim JH, Song HK. Swapping of interaction partners with ATG5 for autophagosome maturation. BMB Rep. 2015; 48:129–130.

31. Song L, Ma L, Chen G, Huang Y, Sun X, Jiang C, Liu H. Autophagy inhibitor 3-methyladenine enhances the sensitivity of nasopharyngeal carcinoma cells to chemotherapy and radiotherapy. J Cent South Univ (Med Sci). 2016; 41:9–18.
32. Zhao Y, Xiong X, Jia L, Sun Y. Targeting Cullin-RING ligases by MLN4924 induces autophagy via modulating the HIF1-REDD1-TSC1-mTORC1-DEPTOR axis. Cell Death Dis. 2012; 3:e386.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download