Abstract
Asthma is considered the hallmark of chronic airway inflammation, in which several inflammatory cells of the innate and adaptive immune system act together. The disease is thought to be caused by a combination of genetic and environmental factors; however, precise mechanisms for airway inflammation remain unclear. The human microbiota provides an increasingly favored explanation for inflammatory diseases; an altered microbiota composition has been shown to regulate immune responses. However, given the complexity of the microbiota, additional research is needed to elucidate its role in the development of disease. One of the candidate molecules that link microbiota to disease is the extracellular vesicles (EVs). EVs are secreted by diverse cell types and they possess the pathophysiological function of delivering signals between bacteria and host. We discuss the role of the microbiota in the development of asthma through releasing EVs.
Asthma is a chronic inflammation characterized by airway hyperresponsiveness, airway narrowing, mucus overproduction, and airway remodeling. Airway inflammation via secretion of pro-inflammatory cytokines is essential for asthma development. These cytokines orchestrate allergic immune responses including T cell differentiation, IgE production, and leukocyte activation.12 Traditionally, allergic inflammation has been considered the result of environmental allergens such as dust mites, pollens, and animal dander. However, recent studies propose that bacteria-derived toxins or components can also be involved in allergic responses. Little is known about the mechanism by which environmental microbiota might contribute to allergic reactions.
A recent metagenomic data analysis revealed a microbiome in the respiratory tract, which was once believed to be sterile, mainly because of difficulties in culturing of the bacteria.3 Yet the airway is external to the body, and this space is thus a primary site for exposure to environmental factors such as bacteria and viruses. Differing compositions of lung microbiota between asthmatic and healthy people imply that bacteria might contribute to the development of disease.45 It is suggested that the local microbiota can directly affect airway-resident cells. However, the microbiota residing at other sites such as the gut might also affect lung immune responses. Further studies might elucidate interactions between the microbiota and the immune system that contribute to asthma.
An effective immune response requires appropriate cytokines and other inflammatory mediators produced by infected or resident cells. Microbiota- and host-derived molecules such as extracellular vesicles (EVs) might also regulate inflammation. EVs are lipid bilayers containing transmembrane proteins, cytosolic proteins, lipids, and nucleic acids with a diverse range of sizes (100–1,000 nm in diameter). As these molecules have surface ligands that interact with receptors on target cells, they can attach and modify the physiological state of recipient cells.67 Recent studies have demonstrated that EVs are involved in development of cancer, atherosclerosis, diabetes, meningitis, and salpingitis.
This review focuses on the potential relationship between environmental microbiota and asthma. We are of the view that asthma is not simply induced by allergens. We also propose that EVs are key molecules that link the microbiota to asthma by regulating immune responses.
Asthma is a worldwide disease commonly characterized by eosinophilic inflammation. Given the large number of eosinophils in the airways, asthma is considered a hallmark of T helper type 2 (Th2) disorders of the lungs. The Th2 response is generally driven by the cytokines IL-4, IL-5, IL-9, and IL-13, which promote increased numbers of eosinophils in the airway and lead to high IgE levels in the blood. In mice, depletion of Th2-associated cytokines or induction of the Th1 response has reduced asthma features.89 However, there is an increasing interest in understanding the non-Th2 response, which likely represents a large proportion of asthma cases.
The non-Th2 response involves adult-onset and severe asthma, which have a mixed Th1 and Th17 response with neutrophilic inflammation.1011 In mice models of asthma, IL-17 has been strongly linked to neutrophilic inflammation, and has led to corticosteroid resistance.1213 Severe asthma driven by IL-17 pathologically correlated with steroid-resistant asthma. Neutrophilic asthma has been linked to upregulation in TNF-α pathways, but it remains uncertain whether TNF blockade could improve steroid responsiveness.1415 Although 2 forms of asthma have been defined, Th2 and non-Th2 responses co-occur in many cases, rather than being mutually exclusive.
Similar to many chronic inflammatory diseases, the pathogenesis of asthma mainly depends on genetic susceptibility and environmental factors.161718 Environmental changes are a major factor in the development of allergic diseases. The recent microbiota hypothesis has suggested that perturbations in the gut microbiota disrupt the mucosal tolerance.1920 A balance between the microbiota and host that maintains functional homeostasis is accurately regulated; however, it is constantly challenged by several factors. Once pathogenic bacteria colonize a niche, they cause a state of dysbiosis in the microbial community, which is now recognized to induce allergic diseases such as asthma (Table).
The presence of certain bacteria in the airway can regulate allergic inflammation.21 Haemophilus influenzae might be related to neutrophilic asthma, in which inflammation is mainly induced by neutrophils.22 Another well-known pathogenic bacterium, Staphylococcus aureus, is thought to be involved in severe asthma, since asthmatic patients have presented high levels of specific antibodies against S. aureus enterotoxins.2324 Other bacteria such as Moraxella catarrhalis and Streptococcus pneumoniae have also been associated with childhood asthma.25 However, not all bacteria in the airway aggravate the severity of asthma; for example, Lactobacillus rhamnosus has protective effects against respiratory infection.26 Specific bacteria seem to modulate allergic inflammation; however, whether a single bacterial species can induce allergic disease is still unclear.
Recent studies have revealed that the composition of the microbiota residing in the lung can be more important in allergic diseases than the simple presence of individual species. Analyses of 16S rRNA have found that the airway hosts a complex community of microbes; moreover, bacterial populations of asthmatic patients are unlike those of healthy control subjects.2728 The bacterial compositions of bronchial samples have also exhibited differences between 2 groups.2930 These reports indicate that the phylum Proteobacteria was relatively prevalent in asthmatics, while phylum Bacteroidetes predominated in healthy controls. It has become clear that microbial community dysbiosis correlates with asthma.3132 Diverse bacterial communities reside in the airway, and altered composition of these microbiota might contribute to allergic inflammation in asthma.
Clinical studies of corticosteroid inhalation suggest that the airway microbiota affects corticosteroid responsiveness among asthmatic patients.33 Bacterial community profiles of corticosteroid-sensitive or -resistant asthmatics were not well discerned, but Haemophilus parainfluenzae was observed in some of the corticosteroid-resistant asthmatics. Another study of antibiotic treatment observed azithromycin-induced modification of the airway microbiota of adult asthmatic patients.34 The abundance of members of the Haemophilus and Pseudomonas genera declined within the community, but the abundance of Anaerococcus species increased. These findings suggest that the airway microbiota drives asthma development, and explain corticosteroid responsiveness. We expect that current efforts to understand the airway microbiome membership in detail will help reveal a mechanistic link between the microbiota and asthma.
The airway microbiota can shape lung-specific immune responses resulting in either homeostatic or detrimental inflammation. Recently, the gastrointestinal (GI) tract microbiome has also become a plausible influencer of immune development against allergy or asthma. This interest in GI microbes stemmed from an observed inverse relationship between rate of childhood asthma and exposure to bacteria in infancy.35363738 Moreover, infants who develop asthma have presented relatively few lactobacillus, and bifidobacteria, and relatively many enterococcus.394041 Further studies indicate that the gut microbiota influences the maturation of immune function in early life through the oral ingestion of environmental bacteria.424344
Studies in mice also provide strong evidence of the role of gut microbiota in the regulation of immune function. Germ-free (GF) mice presented strong allergic inflammation compared to specific pathogen-free (SPF) mice through OVA sensitization and challenge.45 Specific bacteria like Lactobacillus reuteri exhibited the protective effects of inducing regulatory T (Treg) cell expansion and reducing inflammation, in response to OVA challenge in sensitized BALB/c mice.46 Treatment with Clostridium strains produced similar effects on Treg cells in the colonic mucosa, and reduced IgE levels after OVA sensitization.47 Interestingly, segmented filamentous bacteria induced Th17 cell expansion, which mediates mucosal defense in the lamina propria.48 However, a recent study found that direct ingestion of certain bacteria was insufficient to induce significant changes in immune function.
Experimental feeding of different dietary components indicated that dietary changes could affect allergic inflammation by modifying the microbiota composition. High fiber diet altered the ratio of Firmicutes spp. to Bacteroidetes spp., and increased the levels of circulating short-chain fatty acids. These subsequent alterations protected against allergic lung inflammation via activation of Treg cells.4950 The mechanisms of Treg-mediated suppression are not fully understood, but microbes do promote Treg cell activation by maintaining resident dendritic cells (DCs) in an immature state, which is essential for mucosal tolerance.5152
The propensity of an innocuous antigen to induce local and systemic immune unresponsiveness is termed as oral tolerance.53 In the large intestine, commensal bacteria are also regulated by an analogous, but more locally processed, tolerance. It is assumed that oral tolerance and airway tolerance are tightly linked, and that the GI tract might act as a sensor for the development of tolerance to antigens. However, it remains to be determined how changes in the gut microbiota affect lung immunity.
Immune system recognition of pathogens is essential to activating immune cells. Bacterial components that trigger immune response are often referred to as pathogen-associated molecular patterns (PAMPs); these components include carbohydrates, lipids, proteins, and genetic material. PAMPs can bind to pattern recognition receptors (PRRs) on immune and non-immune cells, thereby initiating signaling cascades. Given their role in the immune system, pathogens have evolved to increase their virulence by modulating their signals to PRRs.54 Recent studies report that both PAMPs and EVs released from infected cells and pathogens are likely to be involved in immune responses.55
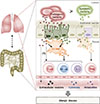
EVs are released by diverse cell types that have been implicated in allergic responses such as bronchial epithelial cells, mast cells, dendritic cells, and T cells in the lung.56 Bronchial epithelial cells are the primary producers of EVs in the lungs of patients with asthma.57 Interestingly, it was found that IL-13-stimulated bronchial epithelial cells released EVs that promoted proliferation of macrophages, but reduction in EV secretion seemed to ameliorate asthmatic features. During allergic inflammation, mast cell-derived EVs were highly activated, and contributed to induction of DC maturation.58 DCs also produce EVs that present allergens and activate allergen-specific Th2 cells.59 Many studies suggest that EVs can transfer MHC/antigen complexes, enabling the DCs to efficiently activate T cells.6061 These observations indicate the potential contributions of diversely sourced EVs to the pathogenesis of asthma. In contrast, some studies have reported that EVs can inhibit Th2 response-associated cytokine production, IgE response, and even prevent the development of asthma.62 While immune cells mediate immune responses, non-immune cells such as bacteria also have important roles in activating immune responses. EVs produced by pathogens such as Staphylococcus aureus have been implicated in atopic dermatitis-like skin inflammation;6364 these EVs might also induce neutrophilic pulmonary inflammation.65 EVs derived from E. coli induce several infectious diseases, and can aggravate emphysema through IL-17A-mediated neutrophilic inflammation.66 Pseudomonas aeruginosa often contributes to lung diseases such as cystic fibrosis, and its EVs increase pulmonary inflammation through Toll-like receptor (TLR)2 and TLR4 pathways.67 Moreover, indoor dust, which contains many components of bacteria, has been associated with both Th1 and Th17 responses, which induce neutrophilic pulmonary inflammation.6869 As bacteria-derived EVs can affect distal host cell sites,70 these vesicles might play significant roles throughout the body. Here, we propose a model that uses EVs to link the environmental microbiota with airway immunity (Figure).
The presence of lung and gut dysbiosis in asthma suggests that microbiota composition might shape the immune system and lead to the development of asthma. Microbial communities have immunomodulatory roles in both the disease progression and the clinical outcome of asthma, but our understanding of mechanisms linking gut- to lung-immunity is still unclear. EVs might provide new insights into immune responses, as they can move to distant sites and stimulate other cells such as immune and epithelial cells. EVs produced by commensal bacteria can benefit the host by promoting mucosal tolerance and protecting against the onset of diseases. However, questions remain as to which components of EVs activate recipient cells and how EVs regulate intracellular signaling pathways. Further studies will provide a mechanistic understanding of interactions between the microbiota and the host immune response through EVs, which in turn will aid in providing new insights into the management of asthma.
Figures and Tables
Figure
Relationship between lung immunity and gut immunity. Extracellular vesicles (EVs) produced by lung and gut microbes can activate epithelial and immune cells. These cells also secrete EVs with immunological functions.
Table
Pathogenic and beneficial bacteria associated with allergic inflammation
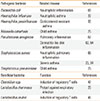
| Pathogenic bacteria | Related disease | References |
|---|---|---|
| Escherichia coli | Neutrophilic inflammation | 66 |
| Haemophilus influenzae | Neutrophilic asthma | 22 |
| Haemophilus parainfluenzae | Corticosteroid-resistant asthma | 33 |
| Moraxella catarrhalis | Child asthma | 25 |
| Pseudomonas aeruginosa | Pulmonary inflammation | 67 |
| Dermatitis-like skin inflammation | 63, 64 | |
| Staphylococcus aureus | Neutrophilic pulmonary inflammation | 65 |
| Severe asthma | 23, 24 | |
| Streptococcus pneumoniase | Child asthma | 25 |
| Beneficial bacteria | Function | References |
|---|---|---|
| Clostridium spp. | Induction of regulatory T cells | 47 |
| Lactobacillus rhamnosus | Protect against respiratory infection | 26 |
| Lactobacillus reuteri | Induction of regulatory T cells | 46 |
ACKNOWLEDGMENTS
This study was supported by a grant from the Korean Health Technology R&D Project, Ministry of Health & Welfare, ROK (HI14C2628).
References
1. Cohn L, Elias JA, Chupp GL. Asthma: mechanisms of disease persistence and progression. Annu Rev Immunol. 2004; 22:789–815.
2. Wills-Karp M. Immunologic basis of antigen-induced airway hyperresponsiveness. Annu Rev Immunol. 1999; 17:255–281.
3. Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999; 65:4799–4807.
4. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010; 5:e8578.
5. Charlson ES, Bittinger K, Chen J, Diamond JM, Li H, Collman RG, et al. Assessing bacterial populations in the lung by replicate analysis of samples from the upper and lower respiratory tracts. PLoS One. 2012; 7:e42786.
6. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013; 200:373–383.
7. Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014; 30:255–289.
8. Brusselle GG, Kips JC, Tavernier JH, van der Heyden JG, Cuvelier CA, Pauwels RA, et al. Attenuation of allergic airway inflammation in IL-4 deficient mice. Clin Exp Allergy. 1994; 24:73–80.
9. Gavett SH, O'Hearn DJ, Li X, Huang SK, Finkelman FD, Wills-Karp M. Interleukin 12 inhibits antigen-induced airway hyperresponsiveness, inflammation, and Th2 cytokine expression in mice. J Exp Med. 1995; 182:1527–1536.
10. Shaw DE, Berry MA, Hargadon B, McKenna S, Shelley MJ, Green RH, et al. Association between neutrophilic airway inflammation and airflow limitation in adults with asthma. Chest. 2007; 132:1871–1875.
11. Manni ML, Trudeau JB, Scheller EV, Mandalapu S, Elloso MM, Kolls JK, et al. The complex relationship between inflammation and lung function in severe asthma. Mucosal Immunol. 2014; 7:1186–1198.
12. McKinley L, Alcorn JF, Peterson A, Dupont RB, Kapadia S, Logar A, et al. TH17 cells mediate steroid-resistant airway inflammation and airway hyperresponsiveness in mice. J Immunol. 2008; 181:4089–4097.
13. Lajoie S, Lewkowich IP, Suzuki Y, Clark JR, Sproles AA, Dienger K, et al. Complement-mediated regulation of the IL-17A axis is a central genetic determinant of the severity of experimental allergic asthma. Nat Immunol. 2010; 11:928–935.
14. Berry MA, Hargadon B, Shelley M, Parker D, Shaw DE, Green RH, et al. Evidence of a role of tumor necrosis factor alpha in refractory asthma. N Engl J Med. 2006; 354:697–708.
15. Wenzel SE, Barnes PJ, Bleecker ER, Bousquet J, Busse W, Dahlen SE, et al. A randomized, double-blind, placebo-controlled study of tumor necrosis factor-alpha blockade in severe persistent asthma. Am J Respir Crit Care Med. 2009; 179:549–558.
16. Chen Y, Wong GW, Li J. Environmental Exposure and Genetic Predisposition as Risk Factors for Asthma in China. Allergy Asthma Immunol Res. 2016; 8:92–100.
17. Anderson GP. Endotyping asthma: new insights into key pathogenic mechanisms in a complex, heterogeneous disease. Lancet. 2008; 372:1107–1119.
18. Kim BJ, Lee SY, Kim HB, Lee E, Hong SJ. Environmental changes, microbiota, and allergic diseases. Allergy Asthma Immunol Res. 2014; 6:389–400.
19. Noverr MC, Huffnagle GB. The ‘microflora hypothesis’ of allergic diseases. Clin Exp Allergy. 2005; 35:1511–1520.
20. Blaser MJ. Who are we? Indigenous microbes and the ecology of human diseases. EMBO Rep. 2006; 7:956–960.
21. Gollwitzer ES, Saglani S, Trompette A, Yadava K, Sherburn R, McCoy KD, et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med. 2014; 20:642–647.
22. Essilfie AT, Simpson JL, Dunkley ML, Morgan LC, Oliver BG, Gibson PG, et al. Combined Haemophilus influenzae respiratory infection and allergic airways disease drives chronic infection and features of neutrophilic asthma. Thorax. 2012; 67:588–599.
23. Bachert C, Gevaert P, Howarth P, Holtappels G, van Cauwenberge P, Johansson SG. IgE to Staphylococcus aureus enterotoxins in serum is related to severity of asthma. J Allergy Clin Immunol. 2003; 111:1131–1132.
24. Bachert C, van Steen K, Zhang N, Holtappels G, Cattaert T, Maus B, et al. Specific IgE against Staphylococcus aureus enterotoxins: an independent risk factor for asthma. J Allergy Clin Immunol. 2012; 130:376–381.e8.
25. Bisgaard H, Hermansen MN, Buchvald F, Loland L, Halkjaer LB, Bonnelykke K, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med. 2007; 357:1487–1495.
26. Tomosada Y, Chiba E, Zelaya H, Takahashi T, Tsukida K, Kitazawa H, et al. Nasally administered Lactobacillus rhamnosus strains differentially modulate respiratory antiviral immune responses and induce protection against respiratory syncytial virus infection. BMC Immunol. 2013; 14:40.
27. Huang YJ, Nariya S, Harris JM, Lynch SV, Choy DF, Arron JR, et al. The airway microbiome in patients with severe asthma: Associations with disease features and severity. J Allergy Clin Immunol. 2015; 136:874–884.
28. Zhang Q, Cox M, Liang Z, Brinkmann F, Cardenas PA, Duff R, et al. Airway Microbiota in Severe Asthma and Relationship to Asthma Severity and Phenotypes. PLoS One. 2016; 11:e0152724.
29. Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011; 127:372–381.e1-3.
30. Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD. Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol. 2013; 131:346–352.e1-3.
31. Dulek DE, Peebles RS. Bacteria and asthma: more there than we thought. Expert Rev Respir Med. 2011; 5:329–332.
32. Huang YJ, Lynch SV. The emerging relationship between the airway microbiota and chronic respiratory disease: clinical implications. Expert Rev Respir Med. 2011; 5:809–821.
33. Goleva E, Jackson LP, Harris JK, Robertson CE, Sutherland ER, Hall CF, et al. The effects of airway microbiome on corticosteroid responsiveness in asthma. Am J Respir Crit Care Med. 2013; 188:1193–1201.
34. Wong EH, Porter JD, Edwards MR, Johnston SL. The role of macrolides in asthma: current evidence and future directions. Lancet Respir Med. 2014; 2:657–670.
35. Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrlander C, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011; 364:701–709.
36. Heederik D, von Mutius E. Does diversity of environmental microbial exposure matter for the occurrence of allergy and asthma? J Allergy Clin Immunol. 2012; 130:44–50.
37. Riedler J, Braun-Fahrlander C, Eder W, Schreuer M, Waser M, Maisch S, et al. Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet. 2001; 358:1129–1133.
38. Waser M, Michels KB, Bieli C, Floistrup H, Pershagen G, von Mutius E, et al. Inverse association of farm milk consumption with asthma and allergy in rural and suburban populations across Europe. Clin Exp Allergy. 2007; 37:661–670.
39. Bjorksten B, Sepp E, Julge K, Voor T, Mikelsaar M. Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol. 2001; 108:516–520.
40. Kalliomaki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001; 107:129–134.
41. Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007; 56:661–667.
42. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012; 336:489–493.
43. Rautava S, Luoto R, Salminen S, Isolauri E. Microbial contact during pregnancy, intestinal colonization and human disease. Nat Rev Gastroenterol Hepatol. 2012; 9:565–576.
44. Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012; 13:440–447.
45. Herbst T, Sichelstiel A, Schar C, Yadava K, Burki K, Cahenzli J, et al. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011; 184:198–205.
46. Karimi K, Inman MD, Bienenstock J, Forsythe P. Lactobacillus reuteri-induced regulatory T cells protect against an allergic airway response in mice. Am J Respir Crit Care Med. 2009; 179:186–193.
47. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011; 331:337–341.
48. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009; 139:485–498.
49. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014; 20:159–166.
50. Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. 2015; 6:7320.
51. Belkaid Y. Regulatory T cells and infection: a dangerous necessity. Nat Rev Immunol. 2007; 7:875–888.
52. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008; 453:620–625.
53. Mowat AM. Anatomical basis of tolerance and immunity to intestinal antigens. Nat Rev Immunol. 2003; 3:331–341.
54. Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011; 30:16–34.
55. Bhatnagar S, Shinagawa K, Castellino FJ, Schorey JS. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood. 2007; 110:3234–3244.
56. Pyun BY. Extracellular Vesicle: An Unknown Environmental Factor for Causing Airway Disease. Allergy Asthma Immunol Res. 2016; 8:179–180.
57. Kulshreshtha A, Ahmad T, Agrawal A, Ghosh B. Proinflammatory role of epithelial cell-derived exosomes in allergic airway inflammation. J Allergy Clin Immunol. 2013; 131:1194–1203. 1203.e1–1203.e14.
58. Skokos D, Botros HG, Demeure C, Morin J, Peronet R, Birkenmeier G, et al. Mast cell-derived exosomes induce phenotypic and functional maturation of dendritic cells and elicit specific immune responses in vivo. J Immunol. 2003; 170:3037–3045.
59. Admyre C, Telemo E, Almqvist N, Lotvall J, Lahesmaa R, Scheynius A, et al. Exosomes - nanovesicles with possible roles in allergic inflammation. Allergy. 2008; 63:404–408.
60. Hsu DH, Paz P, Villaflor G, Rivas A, Mehta-Damani A, Angevin E, et al. Exosomes as a tumor vaccine: enhancing potency through direct loading of antigenic peptides. J Immunother. 2003; 26:440–450.
61. Andre F, Chaput N, Schartz NE, Flament C, Aubert N, Bernard J, et al. Exosomes as potent cell-free peptide-based vaccine. I. Dendritic cell-derived exosomes transfer functional MHC class I/peptide complexes to dendritic cells. J Immunol. 2004; 172:2126–2136.
62. Prado N, Marazuela EG, Segura E, Fernandez-Garcia H, Villalba M, Thery C, et al. Exosomes from bronchoalveolar fluid of tolerized mice prevent allergic reaction. J Immunol. 2008; 181:1519–1525.
63. Hong SW, Kim MR, Lee EY, Kim JH, Kim YS, Jeon SG, et al. Extracellular vesicles derived from Staphylococcus aureus induce atopic dermatitis-like skin inflammation. Allergy. 2011; 66:351–359.
64. Hong SW, Choi EB, Min TK, Kim JH, Kim MH, Jeon SG, et al. An important role of alpha-hemolysin in extracellular vesicles on the development of atopic dermatitis induced by Staphylococcus aureus. PLoS One. 2014; 9:e100499.
65. Kim MR, Hong SW, Choi EB, Lee WH, Kim YS, Jeon SG, et al. Staphylococcus aureus-derived extracellular vesicles induce neutrophilic pulmonary inflammation via both Th1 and Th17 cell responses. Allergy. 2012; 67:1271–1281.
66. Kim YS, Lee WH, Choi EJ, Choi JP, Heo YJ, Gho YS, et al. Extracellular vesicles derived from Gram-negative bacteria, such as Escherichia coli, induce emphysema mainly via IL-17A-mediated neutrophilic inflammation. J Immunol. 2015; 194:3361–3368.
67. Park KS, Lee J, Jang SC, Kim SR, Jang MH, Lotvall J, et al. Pulmonary inflammation induced by bacteria-free outer membrane vesicles from Pseudomonas aeruginosa. Am J Respir Cell Mol Biol. 2013; 49:637–645.
68. Kim YS, Choi EJ, Lee WH, Choi SJ, Roh TY, Park J, et al. Extracellular vesicles, especially derived from Gram-negative bacteria, in indoor dust induce neutrophilic pulmonary inflammation associated with both Th1 and Th17 cell responses. Clin Exp Allergy. 2013; 43:443–454.
69. Kim YS, Choi JP, Kim MH, Park HK, Yang S, Kim YS, et al. IgG Sensitization to Extracellular Vesicles in Indoor Dust Is Closely Associated With the Prevalence of Non-Eosinophilic Asthma, COPD, and Lung Cancer. Allergy Asthma Immunol Res. 2016; 8:198–205.
70. Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev. 2010; 74:81–94.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download