Abstract
Echovirus 30 is one of the distinct serotypes of enteroviruses and commonly isolated agent causing sporadic to large outbreaks with aseptic meningitis in many regions over the world. Recently, an outbreak of echovirus 30 associated with aseptic meningitis occurred in Korea in 2008. In order to analyze echovirus 30 in Korea, the virus was isolated from cerebrospinal fluid samples of a male patient with aseptic meningitis and its genome sequence was determined. The sequence of Korean echovirus 30 isolate was compared with those of reference strains (Bastianni, FDJS03-84, zhejiang-17-03, 14916net87). At the nucleotide level, the P1 region (84.8∼89.0%) had the highest identity value; at the amino acid level, the P3 region (97.0∼98.5%) showed the highest value. When the cleavage sites were compared, most sites were identical except those between VP1 and 2A; the Bastianni stain had TT/GA, whereas the other four strains contained NT/GA. The China strains (FDJS03-84 and zhejiang-17-03) were grouped together and the other strains were distinct from each branch in the phylogenetic tree based on the complete genome sequences.
REFERENCES
1). Huttunen P, Santti J, Pulli T, Hyypiä T. The major echovirus group is genetically coherent and related to coxsackie B viruses. J Gen Virol. 1996; 77:715–25.

2). Mayo MA, Pringle CR. Virus taxonomy. J Gen Virol. 1998; 79:649–57.
3). Oberste MS, Maher K, Kilpatrick DR, Pallansch MA. Molecular evolution of the human enteroviruses: Correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol. 1999; 73:1941–8.

4). Kok PW, Leeuwenburg J, Tukei P, van Wezel AL, Kapsenberg JG, van Steenis G, et al. Serological and virological assessment of oral and inactivated poliovirus vaccines in a rural population in Kenya. Bull World Health Organ. 1992; 70:93–103.
5). Yerly S, Gervaix A, Simonet V, Caflisch M, Perrin L, Wunderli W. Rapid and sensitive detection of enteroviruses in specimens from patients with aseptic meningitis. J Clin Microbiol. 1996; 34:199–201.

6). Baek K, Yeo S, Lee B, Park K, Song J, Yu J, et al. Epidemics of enterovirus infection in Chungnam Korea, 2008 and 2009. Virol J. 2011; 8:297.

7). Jee YM, Cheon DS, Choi WY, Ahn JB, Kim KS, Chung YS, et al. Updates on enterovirus surveillance in Korea. Inf Chemotherapy. 2004; 36:294–303.
8). Racaniello VR, Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci U S A. 1981; 78:4887–91.

9). Pettersson RF, Ambros V, Baltimore D. Identification of a protein linked to nascent poliovirus RNA and to the polyuridylic acid of negative-strand RNA. J Virol. 1978; 27:357–65.

10). Park K, Song J, Baek K, Lee C, Kim D, Cho S, et al. Genetic diversity of a Korean echovirus 5 isolate and response of the strain to five antiviral drugs. Virol J. 2011; 8:79.

11). Park K, Yeo S, Baek K, Cheon D, Choi Y, Park J, et al. Molecular characterization and antiviral activity test of common drugs against echovirus 18 isolated in Korea. Virol J. 2011; 8:516.

12). Lindberg AM, Andersson P, Savolainen C, Mulders MN, Hovi T. Evolution of the genome of Human enterovirus B: incongruence between phylogenies of the VP1 and 3CD regions indicates frequent recombination within the species. J Gen Virol. 2003; 84:1223–35.

13). Paananen A, Savolainen-Kopra C, Kaijalainen S, Vaarala O, Hovi T, Roivainen M. Genetic and phenotypic diversity of echovirus 30 strains and pathogenesis of type 1 diabetes. J Med Virol. 2007; 79:945–55.

14). Zhao YN, Perlin DS, Park S, Jiang RJ, Chen L, Chen Y, et al. FDJS03 isolates causing an outbreak of aseptic meningitis in China that evolved from a distinct Echovirus 30 lineage imported from countries of the Commonwealth of Independent States. J Clin Microbiol. 2006; 44:4142–8.

15). Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987; 4:406–25.
16). Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22:4673–80.

17). Drake JW. Rates of spontaneous mutation among RNA viruses. Proc Natl Acad Sci U S A. 1993; 90:4171–5.

18). Guillot S, Caro V, Cuervo N, Korotkova E, Combiescu M, Persu A, et al. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J Virol. 2000; 74:8434–43.

19). Santti J, Hyypiä T, Kinnunen L, Salminen M. Evidence of recombination among enteroviruses. J Virol. 1999; 73:8741–9.

20). Wang HY, Xu AQ, Zhu Z, Li Y, Ji F, Zhang Y, et al. The genetic characterization and molecular evolution of echovirus 30 during outbreaks of aseptic meningitis. Zhonghua Liu Xing Bing Xue Za Zhi. 2006; 27:793–7.
21). Wang JR, Tsai HP, Huang SW, Kuo PH, Kiang D, Liu CC. Laboratory diagnosis and genetic analysis of an echovirus 30-associated outbreak of aseptic meningitis in Taiwan in 2001. J Clin Microbiol. 2002; 40:4439–44.

22). Zhao YN, Jiang QW, Jiang RJ, Chen L, Perlin DS. Echovirus 30, Jiangsu Province, China. Emerg Infect Dis. 2005; 11:562–7.

23). Chen GW, Huang JH, Lo YL, Tsao KC, Chang SC. Mosaic genome structure of echovirus type 30 that circulated in Taiwan in 2001. Arch Virol. 2007; 152:1807–17.

24). Paananen A, Savolainen-Kopra C, Kaijalainen S, Vaarala O, Hovi T, Roivainen M. Genetic and phenotypic diversity of echovirus 30 strains and pathogenesis of type 1 diabetes. J Med Virol. 2007; 79:945–55.

25). Zhao YN, Perlin DS, Park S, Jiang RJ, Chen L, Chen Y, et al. FDJS03 isolates causing an outbreak of aseptic meningitis in China that evolved from a distinct Echovirus 30 lineage imported from countries of the Commonwealth of Independent States. J Clin Microbiol. 2006; 44:4142–8.

26). Muir P, Kämmerer U, Korn K, Mulders MN, Pöyry T, Weissbrich B, et al. Molecular typing of enteroviruses: current status and future requirements. The European Union Concerted Action on Virus Meningitis and Encephalitis. Clin Microbiol Rev. 1998; 11:202–27.
27). Kinnunen L, Pöyry T, Hovi T. Genetic diversity and rapid evolution of poliovirus in human hosts. Curr Top Microbiol Immunol. 1992; 176:49–61.

28). Diedrich S, Driesel G, Schreier E. Sequence comparison of echovirus type 30 isolates to other enteroviruses in the 5′ noncoding region. J Med Virol. 1995; 46:148–52.
Figure 1.
Phylogenetic analysis based on the complete genome sequence of Korean echovirus 30 and reference strains. Nucleotide sequences were analyzed using the neighbor-joining method.
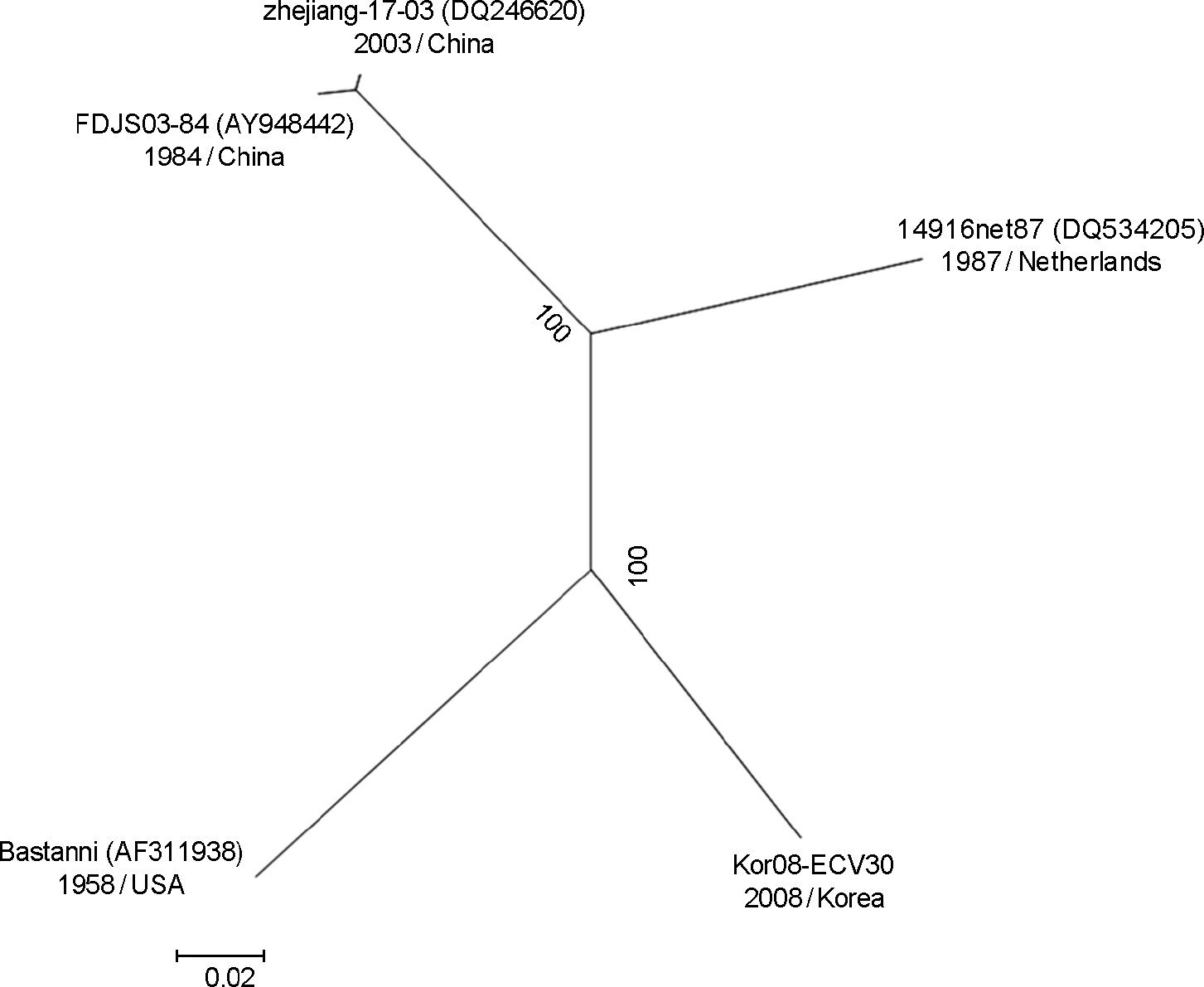
Table 1.
Specific primer sets for PCR and sequencing of Echovirus 30
Table 2.
Genome components of Korean Echovirus 30 isolate
Table 3.
The percentage of identities in nucleotide and amino acid sequence between the Korean Echovirus 30 and Bastianni strain
Table 4.
Predicted N-terminal cleavage sites of Korean Echovirus 30 and reference strains




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download