Abstract
Fowl adenovirus (FAdV) is distributed worldwide and causes economic losses in the poultry industry. The objectives of this study were to determine the hexon and fiber gene changes in an attenuated FAdV isolate from Malaysia in specific pathogen-free chicken embryonated eggs (SPF CEE) and its infectivity in commercial broiler chickens. SPF CEE were inoculated with 0.1 mL FAdV inoculum via the chorioallantoic membrane (CAM) for 20 consecutive passages. The isolate at passage 20 (E20), with a virus titer of 108.7TCID50/mL (TCID50, 50% tissue culture infective dose), was inoculated (0.5 mL) into one-day-old commercial broiler chicks either via oral or intraperitoneal routes. The study demonstrated that 100% embryonic mortality was recorded from E2 to E20 with a delayed pattern at E17 onwards. The lesions were confined to the liver and CAM. Substitutions of amino acids in the L1 loop of hexon at positions 49 and 66, and in the knob of fiber at positions 318 and 322 were recorded in the E20 isolate. The isolate belongs to serotype 8b and is non-pathogenic to broiler chickens, but it is able to induce a FAdV antibody titer. It appears that molecular changes in the L1 loop of hexon and the knob of fiber are markers for FAdV infectivity.
Fowl adenovirus (FAdV) is a double-stranded DNA virus with a non-enveloped structure and is comprised of 12 serotypes among five molecular species designated by letters A to E [4]. Highly pathogenic FAdV is the main causative agent of inclusion body hepatitis (IBH), hydropericardium syndrome, and gizzard erosion in chickens [516]. To date, serotype 8b and 11 are the most common FAdV isolates causing IBH outbreaks with serious economic impact on the poultry industry worldwide [111621]. IBH outbreaks in commercial broiler chickens in Malaysia continue to increase, and a vaccine against IBH is not available [81017]. Embryonated chicken eggs or cell cultures have been commonly used as an alternative host for FAdV propagation and attenuation [14].
Molecular analysis of FAdV shows that the major structural proteins encoded for antigenic specificity are frequently identified in the hexon and fiber genes sequences [615]. However, the roles of virulence determinants in hexon and fiber proteins remain obscure although it was speculated that the fiber gene encoded FAdV infectivity [618]. It was reported that molecular changes in the L1 loop of a propagated FAdV isolate resulted in attenuation; however, the position of the nucleotide and amino acid changes were unknown [14]. In addition, there has been little reported on the effect of serial passages of FAdV isolate in embryonated chicken eggs in elucidating the molecular changes in the fiber gene that may affect viral infectivity. Furthermore, markers gene for FAdV attenuation remain unknown following passages in alternative host. It was the objectives of this study to determine the hexon and fiber gene changes in an attenuated FAdV isolate from Malaysia (UPM1137). The changes were identified in specific pathogen-free chicken embryonated eggs (SPF CEE) at passage 20 (E20), and the infectivity of the isolate in commercial broiler chickens was assessed.
The FAdV isolate UPM1137 was obtained from 27-week-old commercial layer chickens during a 2011 IBH outbreak on a farm with 2% mortality. The livers of the affected chickens were pale and friable, and gizzard erosion at the koilin layer was also observed. Virus inoculum from liver samples was prepared based on methods described previously [1]. Briefly, the liver was homogenized in suspension of sterile phosphate buffered saline (PBS; pH 7.4, 0.1 M) followed by centrifugation at 381 × g for 30 min and purified by using a 0.45 µm syringe filter (Sartorius, Germany). Antibiotic-antimycotic solution (Gibco, USA) was added to the liver inoculum at a dilution of 1 in 10 (v/v) and the mixture incubated for 1 h at 4℃ prior to inoculation into SPF CEE.
Nine-day-old SPF CEE were inoculated with 0.1 mL FAdV inoculum via the chorioallantoic membrane (CAM) route and candled daily to detect mortality [1]. Mortality and the presence of gross and histological lesions in the dead embryos were recorded throughout the passages. Liver and CAM were harvested and placed in 10% buffered formalin for histological examination and stained with H&E [3]. For subsequent passages (up to 20 consecutive passages), the harvested liver was processed and used as the inoculum.
At E20, the FAdV isolate was titrated (50% tissue culture infective dose, TCID50) using primary chicken embryo liver (CEL) cells. Briefly, a 96-well tissue culture plate (TPP, Switzerland) seeded with a monolayer of CEL cells and inoculated with 0.1 mL of inoculum from E20 after dilution into 10-fold serial dilution (10−1 to 10−8). Each well was monitored daily for cytopathic effects, and the titer was determined based on methods described by Reed and Muench [20].
The liver homogenates at E2, E5, E10, E15, and E20 were extracted for DNA by using i-Genomic DNA Extraction Mini kit (iNtRON, Korea) according to the manufacturer's recommendations. DNA concentration was measured by biophotometer (Eppendorf, Germany) at 260/280 nm wavelengths. Amplification of DNA for detection of FAdV was conducted by using a PCR master mix (Bioline, UK) according to the manufacturer's protocol. Two set of published primers based on the hexon gene, H1/H2 and H3/H4 [19], and a newly designed fiber gene primer, FibF/FibR (FibF: 5′-GGTCTA CCCCTTTTGGCTCC-3′ and FibR: 5′-GCGTCGTAGATGA AGGGAGG-3′) based on FAdV-8 reference strain [18], were used in this study. The PCR product was analyzed via 1% agarose gel electrophoresis at 70 V for 45 min prior to staining with RedSafe Nuclei Acid Staining solution (iNtRON) and visualized under UV transillumination. All positive products were purified by using a MEGAquick-spin Total Fragment DNA Purification kit (iNtRON) following the recommended protocol. The purified product was cloned into pCR 2.1-TOPO vector using a TOPO TA Cloning kit (Invitrogen, USA) prior plasmid purification by DNA-spin plasmid DNA purification kit (iNtRON).
The plasmid was submitted to a sequencing service (First BASE, Singapore) for at least three sequence determinations using universal primers, T7 promoter, and M13R_pUC26 for both directions in order to obtain consensus sequences. All nucleotide sequences were assembled, edited, and analyzed using BioEdit (ver. 7.2.5; Ibis Therapeutics, USA) and were translated into amino acid sequences by using the online ExPASy tool program (SIB Swiss Institute of Bioinformatics, Switzerland). The E2 FAdV, designated as UPM1137E2 with GenBank accession Nos. KF866370 and KY305950 based on hexon and fiber genes, respectively, was used as the reference for detection of molecular changes in the FAdV isolate at E5, E10, E15, and E20. Detection was performed by using the ClustalW method [12]. The number of nucleotides and amino acids different was determined by using a sequence difference count matrix and the percentage identity was determined via a sequence identity matrix. The position of the L1 loop in the hexon gene was located at residues 101 to 298, corresponding to the HG FAdV strain [7]. For the fiber gene, three different regions were determined (tail, residues 1–75; shaft, residues 76–356; knob, residues 369–523) [6]. All of the nucleotide sequences of the hexon and fiber genes acquired in this study have been submitted to GenBank under accession Nos. KY911365 to KY911368 (hexon) and KY911369 to KY911372 (fiber). Based on the hexon and fiber genes, 30 published reference FAdV strains and their GenBank accession Nos. were retrieved for phylogenetic tree construction. Hexon gene-based strains: CELO (Z67970), PL/060/08 (GU952110), 340 (AF508952), TR22 (AF508953), KR5 (AF508951), B1-7 (KU342001), J2-A (AF339917), C2B (EU979377), SR48 (AF508946), P7-A (AF339915), SR49 (AF508948), 75 (AF508949), A-2A (NC_000899), 380 (KT862812), 1047 (DQ323984), UF71 (EU979378), CR119 (AF508954), YR36 (AF508955), B-3A (AF339922), 58 (AF508957), TR59 (EU979374), 764 (JN112373), Australian FAdV vaccine (GU120268), 430-06 (GU120266), HG (GU734104), UPM08158 (JF917238), UPM08136 (JF917239), UPM04217 (KU517714), Duck adenovirus (KF286430), and Turkey adenovirus (AC_000016). Fiber gene-based strains: CELO (U46933), OTE (FN557186), PL/060/08 (GU952108), 08-3622 (FN557184), 340 (FR872928), B1-7 (KU342001), Bareilly (FJ949088), Kr-Yeoju (HQ709232), Kr-Gunwi (HQ709231), AG234 (HE608153), INT4 (FR872911), ON1 (GU188428), C2B (HE608154), SR48 (LN907576), 685 (KT862805), SR49 (LN907578), A-2A (NC_000899), 380 (LN907574), 05-50052-2924-1 (JQ034217), 05-50052-3181 (JQ034218), CR119 (LN907584), YR36 (LN907582), TR59 (KT037703), 764 (KT037711), HG (GU734104), CFA3 (FAU40588), CFA40 (AF155911), UPM04217 (KU517714), Duck adenovirus (KF286430), and Turkey adenovirus (AC_000016). Amino acid sequences with 198 lengths in the L1 loop of the hexon gene and the entire portion of the fiber gene were entered into a distance matrix using the Jones-Taylor-Thorton model. This was followed by tree formation through the neighbor-joining method with 1,000 bootstrap replicates as deployed in MEGA software (ver. 5) [1022].
Sixty-four, one-day-old commercial broiler chicks were divided into three groups: A, B, and C. Each group was divided further into sacrificed and mortality groups. All chicks in Groups A and B were inoculated with 0.5 mL FAdV isolate UPM1137E20 with a virus titer of 108.7TCID50/mL via oral and intraperitoneal routes, respectively. All chicks in Group C remained uninoculated and formed the control group. All chicks were given feed and water ad libitum and were monitored daily throughout the 3-week post-inoculation (pi) period. Four chicks in each sacrificed group were sacrificed at days 3, 7, 14, and 21 pi by cervical dislocation. At day 0 pi, 4 chicks from the control group were also sacrificed. Body weights and blood samples were collected prior to sacrifice. On necropsy, gross lesions were recorded and samples of trachea, liver, and gizzard were fixed in 10% buffered formalin for histological examination. The FAdV antibody titers in all groups were determined by applying the enzyme-linked immunosorbent assay contained in a commercial kit (BioChek, UK) according to the manufacturer's recommendations. The animal study was conducted under the approval of the Institutional Animal Care and Use Committee of Universiti Putra Malaysia (Animal Use Protocol No. R072/2015).
The antibody titers were statistically analyzed by performing analysis of variance (ANOVA) using IBM SPSS Statistics (ver. 22; IBM, USA) with the significance value set at p < 0.05. Significance was determined further by applying the Tukey's honest significant difference test for multiple comparison analyses.
Embryo mortality of 100% was recorded from E2 to E20 throughout the trial. However, the first passage (E1) only resulted in 14% mortality. All embryos in the control group were alive throughout the trial. Variation in mortality pattern was observed between low (E2–E16) and high (E17 onward) passage levels. From E2 to E16, the first embryo mortality was recorded at days 2 and 3 pi. However, from E17 onward, mortality was delayed until day 4 pi and was delayed to day 10 pi in FAdV-adapted embryos at E20. From E2 to E16, embryo mortality of 100% was recorded at days 7 to 9 pi. In contrast, 100% mortality pattern was delayed to day 11 pi in E16 and E18 embryos, and, at the highest passage level (E20), 100% mortality was delayed until day 12 pi (Table 1).
All embryos in the control group were normal throughout the passages, exhibiting thin and transparent CAM (panel A in Fig. 1) and normal liver (panel B in Fig. 1). In FAdV-inoculated eggs, gross lesions mainly appeared in CAM and liver which was consistent and without variation throughout the passages. All embryos that died between days 2 to 6 pi produced mild lesions with thickening of CAM, as well as pale and swollen liver, both in passages earlier than E17 and in later passages. For late mortalities, embryos that died at days 7 to 12 pi had prominent lesions in CAM with thickening and cloudy membranes (panel C in Fig. 1). In addition, the embryo was congested and the liver was swollen with petechial hemorrhages and multifocal to diffuse areas of necrosis (panel D in Fig. 1).
Histologically, the CAM (panel A in Fig. 2) and liver (panel A in Fig. 3) in the control group were normal throughout the trial. In contrast, mild hepatic degeneration, necrosis, and hemorrhages with basophilic intranuclear inclusion bodies (INIB) were detected in the CAM and liver of inoculated eggs at days 2 to 6 pi. In late mortality embryos, day 7 pi onwards, moderate to severe degeneration, necrosis, and hemorrhages were detected in CAM (panel B in Fig. 2) and liver (panel B in Fig. 3) with numerous basophilic INIB throughout the passages.
The FAdV isolates at E2 (original isolate), E5, E10, E15, and E20 were positive for FAdV with the expected PCR fragment sizes of 1219 bp, 1319 bp, and 1124 bp obtained when using H1/H2, H3/H4, and FibF/FibR primers, respectively (Fig. 4).
The nucleotide sequence of the hexon and fiber genes with lengths of 1166 bp and 1094 bp, respectively, in E2, E5, E10, E15, and E20 isolates showed 99% identity to UPM04217 and to strain 764 under serotype 8b. Analysis based on the L1 loop in the hexon gene revealed 99% identity between E2 and all of the isolates passaged in SPF CEE. Moreover, the sequence identity in the knob region of the fiber gene ranged from 97% to 99%, with the lowest identity at the highest passage (E20).
Several nucleotide and amino acid changes in hexon and fiber genes were detected between E2 and the passaged isolates at each fifth consecutive passage. The location of the L1 loop in the hexon gene was position 6 to 601 in the nucleotide (nt) sequence and 2 to 199 in the amino acid (aa) sequence. Nucleotide base substitution at position 90 (T-C) within the hexon L1 loop was identified in E5 to E20 SPF CEE (Fig. 5). An additional 2 nt bases were substituted in E5 (GenBank accession No. KY911365) at positions 581 (C-T) and 951 (A-C) with 1 aa change in the deduced amino acids at position 317 (T-P) (Fig. 6).
In addition, substitution of other 3 nt bases were noticed in E10 (GenBank accession No. KY911366) SPF CEE at positions 215 (A-G), 531 (G-A), and 733 (T-C) resulting in 2 aa changes at positions 177 (E-K) and 244 (M-T), which were different from the bases in the previous assessed passage (E5). At the subsequent assessed passage, E15 (GenBank accession No. KY911367), the changes in E10 had reverted to the original sequence and only 2 nt base changes were induced at positions 90 (T-C) and 118 (A-G) along with a 1 aa substitution at position 39 (N-S). There were 4 nt base changes at the highest passage, E20 (GenBank accession No. KY911368). The changes at positions 90 (T-C), 147 (A-G), 199 (C-T), and 1134 (A-T) resulted in 3 aa substitutions at deduced amino acid positions 49 (T-A), 66 (A-V), and 378 (M-L).
Analysis of the fiber gene involved the tail, shaft and knob regions at positions 1–66, 67–907, and 946–1094, respectively, for nucleotide sequence analysis and at positions 1–22, 23–303, and 316–364, respectively, for amino acid sequence analysis. Adaptation of FAdV isolate in SPF CEE induced several changes in fiber gene from E5 to E20. Nucleotide base substitution at position 1078 (G-C) within the knob region was observed from E5 (GenBank accession No. KY911369) to E20 (Fig. 7) along with a change in a deduced amino acid at position 360 from alanine (A) to proline (P) (Fig. 8). The isolate at E10 (GenBank accession No. KY911370) revealed an additional 1 nt base change and a 1 aa substitution; positions 985 (T-C) and 329 (F-L), respectively.
Following five consecutive passages, new changes were detected in E15 (GenBank accession No. KY911371) involving 3 nt bases at positions 928 (G-A), 973 (A-G), and 1026 (T-C) resulting in alteration of the deduced amino acids at positions 310 (D-N) and 325 (N-D). Both nucleotide and amino acid changes were prominent in E20 (GenBank accession No. KY911372), in which another 5 nt bases were substituted at positions 879 (A-G), 918 (T-C), 952 (C-G), 964 (A-T), and 1062 (A-C), which were distinct from those at the previous assessed passage (E15). At E20, 2 aa substitutions of deduced amino acids were noticed at positions 318 (P-A) and 322 (N-Y) in the knob region. Molecular changes for the E20 isolate mainly occurred in the knob region of the fiber gene.
Phylogenetic tree analyses revealed the FAdV isolate (UPM1137E2) and the E5 to E20 isolates were closely related to the group E species. Among thirty reference strains, the isolates shared a common ancestor with FAdV-8b strains, UPM04217, 764, 430-06, and Australian FAdV vaccine based on the amino acids in the L1 loop of the hexon gene (Fig. 9) and on the entire fiber gene region (Fig. 10). The highest passage (E20) deviated from UPM1137E2 and other early passages and was closely related to the 430-06 and Australian FAdV vaccine strains based on the hexon gene (Fig. 9).
All chickens were normal and did not exhibit any clinical signs associated with FAdV infection throughout the trial. Grossly, the livers, gizzards, and trachea were normal in all groups from day 3 to day 21 pi. Similarly, histological examination of trachea, liver, and gizzard samples in all groups revealed no abnormal findings.
The antibody titers in the control group at days 0, 3, 7, and 14 pi were 7795 ± 1414, 4776 ± 1170, 8536 ± 1542, and 2445 ± 1472, respectively, and it was not detected at day 21 pi. The antibody titers in Group A (oral inoculation) were 3946 ± 1704, 3742 ± 1946, 3278 ± 1997, and 1700 ± 1332 at days 3, 7, 14, and 21 pi, respectively. In Group B (intraperitoneal inoculation), the titers were 5581 ± 2641, 2168 ± 441, 5746 ± 2034, and 1833 ± 792, at days 3, 7, 14, and 21 pi, respectively. The antibody titer at day 21 pi in the FAdV-inoculated chickens (Groups A and B) were significantly higher (p < 0.05) than that in the control group (Group C). However, there was no significant different (p > 0.05) between inoculated Groups A and B (Fig. 11).
Passaging of the FAdV isolate (UPM1137) in SPF CEE resulted in molecular changes in the hexon and fiber genes of the virus. The virus virulence was reduced with delayed mortality in embryos at the high passage level (E17–E20). These changes are consistent with the molecular changes demonstrated in the regions encoded for virulence determinant [18] in the knob region of the fiber gene as well as in the L1 loop of the hexon gene [14]. At low passages (E2–E16), initial embryo mortality was recorded as early as at days 2 and 3 pi. However, from E17 to E20, initial embryo mortality was delayed to day 4 pi, and in E20 mortality was delayed until day 10 pi. The earliest occurrence of 100% mortality was recorded at day 5 pi in E6, which could be due to the high virulence of the virus in early passages [114]. The change in mortality pattern at E17 onwards indicates that the isolate had begun to reduce its virulence, which is associated with attenuation of the isolate [24]. However, it is interesting to note that the titer of the virus (108.7TCID50/mL) at E20 remained high.
The gross and histological lesions were confined to CAM and liver. More severe lesions were recorded in late embryonic death (day 7 pi onward) with thickening and cloudy of CAM as well as hepatic degeneration, necrosis, and petechial hemorrhage [113]. Basophilic INIBs were abundant in CAM and liver at late mortality (day 7 pi onwards) suggesting the presence of a high level of viral replication in the organs [1913]. In contrast, no visible lesions in SPF CEE were observed following passages of FAdV serotype 4 in CEE [14]. These results show that different FAdV serotypes or isolates vary in their virulence.
Passage of the FAdV isolate in SPF CEE resulted in substitution of nucleotide bases at position 90 from cytosine to thymine in the L1 loop of hexon and in the knob region of fiber at position 1078 (G-C). In addition, there was an amino acid alteration at position 360 (A-P). It appears that these changes are necessary for the isolate to maintain its viability and to allow continuous growth in SPF CEE up to 20 passages since these markers gene were not detected in the original isolate (E2).
The present study revealed that variation in the L1 loop in the hexon protein and the knob region of the fiber gene is associated with virus infectivity. This was clearly demonstrated in passages of the FAdV isolate in SPF CEE, where up to 20 consecutive passages induced virus attenuation through several molecular changes in both nucleotide and amino acids level in the hexon and fiber genes of FAdV. Minimal molecular changes in E5 at the L1 loop and knob regions when compared to E2 (original isolate) were reflected by the changes in isolate virulence and the early embryo mortality pattern. Changes in the L1 loop were minimal in E10 and E15 with only 1 aa substitution, although 2 aa changes were noticed in the knob region. Based on the early mortality pattern, it seems that the amino acids involved did not interfere with virus infectivity in CEE. Differences in infectivity between FAdV isolates were determined by an analysis of the knob region with low amino acid sequence identities [6]. In this study, molecular changes were remarkable at the highest passage (E20) with an additional 2 aa substitutions in the L1 loop and knob regions. These results suggest that changes in the L1 loop at positions 49 (T-A) and 66 (A-V) along with changes in deduced amino acids in the knob region at positions 318 (P-A) and 322 (N-Y) in E20 are markers gene for the virus attenuation observed in SPF CEE since these changes were not detected in earlier passages (E5–E15). Moreover, changes in the knob region, as a primary attachment to the host receptor, affected the mortality pattern at the high passage level, changing from an early to a delayed mortality period. This finding suggests that the knob region encodes a virulence determinant [18].
The L1 loop region in hexon and the entire portion of the fiber gene are useful for FAdV classification into specific group and serotype [15]. Based on phylogenetic tree analysis, the passaged isolates were classified into group E species under serotype 8b based on the hexon and fiber proteins. The E20 passaged isolate deviated from earlier passage isolates and was more closely related to the Australian FAdV vaccine and 430-06 strains in the L1 loop-based tree, which could be an indicator of attenuation of the isolate. This was confirmed as the E20 isolate was non-pathogenic in commercial broiler chickens. Neither abnormal clinical signs nor gross or histological lesions were recorded in the liver, gizzard, and trachea of chickens inoculated with the FAdV E20 isolate. These results are consistent with the delayed embryo mortality pattern results and the molecular changes detected in the L1 loop of the hexon gene and the knob of the fiber gene, which involved 2 aa changes that seem to have an important effect on virus infectivity in chickens [614].
Molecular virus gene changes primarily occur in fiber protein followed by the hexon gene due to fiber structure being located at the outermost part of the capsid for attachment to the host cell's receptor. It seems that changes in the knob region reduced binding affinity toward the host cell's receptors [1823] affecting viral infectivity in both CEEs and commercial broiler chickens. It is interesting to note that the hexon gene is also involved in attenuation with several changes in E20 compared to early passages as shown in a previous study [14].
The high FAdV antibody titer in chicks at day 0 pi (one day old) is due to a maternally derived antibody, possibly resulting from natural exposure to FAdV infection in breeder flocks as the chickens involved in this study were not vaccinated against FAdV [2]. Subsequently, the antibody titer declined significantly and was undetectable in chicks at day 21 pi (21 days old). The study has demonstrated that the FAdV E20 isolate could induce high antibody titer level at day 21 pi in chickens inoculated with FAdV via either oral or intraperitoneal routes.
It was concluded that 20 passages of a FAdVserotype 8b isolate in SPF CEE resulted in molecular changes in the hexon and fiber genes; specifically, substitutions of amino acids in the L1 loop of hexon at positions 49 and 66 and in the knob of fiber at positions 318 and 322. The E20 virus isolate is non-pathogenic, and thus, it is suggested that these molecular changes are markers of FAdV infectivity.
Figures and Tables
Fig. 1
Gross lesions of the chorioallantoic membrane (CAM) and liver in chicken embryos following inoculation with fowl adenovirus (FAdV) isolate, UPM1137. (A) CAM of control embryo with a thin and transparent membrane. (B) Normal liver in a control embryo without significant findings. (C) Gross lesions of CAM with thickening and cloudiness in a FAdV-adapted embryo at day 10 post-inoculation (pi) in passage 7 (E7). (D) Diffuse necrosis and petechial hemorrhages of liver (arrow) in a congested embryo at day 10 pi in E7.
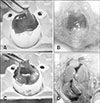
Fig. 2
Histological lesions of chorioallantoic membrane (CAM) following fowl adenovirus inoculation. (A) Normal CAM in control embryos without significant findings. (B) Severe degeneration, necrosis, and hemorrhages in CAM with numerous basophilic intranuclear inclusion bodies (arrows) in embryos at passage 5 at day 8 post-inoculation. H&E stain (A and B). Scale bars = 50 µm (A), 20 µm (B).

Fig. 3
Histological lesions of liver following fowl adenovirus inoculation. (A) Normal liver in control embryos without significant findings. (B) Severe degeneration, necrosis, and hemorrhages in liver with numerous basophilic intranuclear inclusion bodies (arrow) in embryo at passage 20 at day 12 post-inoculation. H&E stain (A and B). Scale bars = 50 µm (A), 20 µm (B).

Fig. 4
Electrophoresis of polymerase chain reaction products using primers H1/H2, H3/H4, and FibF/FibR with fragment sizes of (A) 1219 bp, (B) 1319 bp, and (C) 1124 bp amplifying hexon and fiber gene regions of fowl adenovirus (FAdV) in 1% agarose gel from a sample of the original (passage 2 [E2]) and passaged FAdV isolates in specific pathogen-free chicken embryonated eggs (SPF CEE). Lane M, DNA marker 1 kb; Lane 1, positive control (UPM04217); Lane 2, E2 FAdV; Lane 3, E5 FAdV; Lane 4, E10 FAdV; Lane 5, E15 FAdV; Lane 6, E20 FAdV; Lane 7, negative control.

Fig. 5
Multiple sequence alignment, obtained via ClustalW [12], of 1166 bp nucleotide sequence of the hexon gene between the original fowl adenovirus (FAdV) isolate (UPM1137E2) and passaged FAdV isolates from passages 5 to 20 (E5–E20) in SPF CEE. The position of the L1 loop region is marked in shaded gray from nucleotide base 6 to base 601 with prominent nt substitutions in the E20 isolate.

Fig. 6
Multiple sequence alignment of deduced amino acids in the hexon gene with 388 amino acid residues between the original isolate (UPM1137E2) and passaged fowl adenovirus isolates from passages 5 to 20 (E5–E20) with the L1 loop region (shaded in gray) marked at position 2 to 199. Amino acid substitutions were mainly observed in the variable L1 loop region in the E20 isolate at positions 49 and 66.

Fig. 7
Multiple sequence alignment, obtained by ClustalW [12], of a 1094 bp nucleotide sequence of the fiber gene between the original fowl adenovirus (FAdV) isolate (UPM1137E2) and passaged FAdV isolates from passages 5 to 20 (E5–E20) in SPF CEE. The knob region is shaded in darker gray at positions 946 to 1094, a region with a high number of nucleotide base substitutions mainly in the E20 isolate. The tail of the fiber gene is shaded in very light gray and its shaft is shaded in gray.

Fig. 8
Multiple sequence alignment, obtained by ClustalW [12], of a 364 deduced amino acid sequence of fiber gene between the original fowl adenovirus (FAdV) isolate (UPM1137E2) and passaged FAdV isolates from passage 5 (E5) (UPM1137E5) to E20 (UPM1137E20) with knob region (shaded in darker gray) marked at positions 316 to 364. The tail of fiber is shaded in very light gray and its shaft is shaded in gray. Amino acid substitutions at position 360 were detected from E5 to E20 with an additional two amino acid changes at positions 318 and 322 in the E20 isolate.

Fig. 9
Phylogenetic tree of 198 amino acid residues in the L1 loop of the hexon gene of the fowl adenovirus (FAdV) isolates of this study and of 30 reference FAdV strains retrieved from GenBank. All isolates in SPF CEE from passage 5 (E5) (UPM1137E5) to E20 (UPM1137E20) (shaded in dark gray) were classified as Group E species (shaded in light gray) and were closely related to serotype 8b strains. The early passage isolates E5, E10, and E15 (shaded in dark gray) shared a common ancestor with other Malaysian FAdV strains. The high passage E20 (UPM1137E20) isolate (shaded in dark gray) was closer to the 430-06 and Australian FAdV strains and diverged from the earlier passages isolates. Groups A to E comprised of FAdV strains under serotype 1; serotype 5; serotype 4 and 10; serotype 2, 3, 9, and 11; serotype 6, 7, 8a, and 8b, respectively.

Fig. 10
Phylogenetic tree of fiber gene protein of the fowl adenovirus (FAdV) in this study and 30 reference strains retrieved from GenBank based on 364 amino acid residues. Passaged FAdV isolates, passage 5 (E5) (UPM1137E5), E10 (UPM1137E10), E15 (UPM1137E15), and E20 (UPM1137E20) (shaded in dark gray) were classified under group E species (shaded in light gray) and were closely related to serotype 8b strains. Groups A to E comprised of FAdV strains under serotype 1; serotype 5; serotype 4 and 10; serotype 2, 3, 9, and 11; serotype 6, 7, 8a, and 8b, respectively.

Fig. 11
Fowl adenovirus (FAdV) antibody response in commercial broiler chickens between Group A (oral inoculation), Group B (intraperitoneal inoculation), and Group C (control) from day 0 to 21 post-inoculation (pi) following inoculation with the attenuated FAdV passage 20 (E20) isolate, UPM1137E20. The antibody titer of FAdV was significantly increased in Groups A and B compared to that in the control group at day 21 pi (*p < 0.05). Bars indicate SEM. ELISA, enzyme-linked immunosorbent assay.

Acknowledgments
This study was supported by the Ministry of Higher Education and the Ministry of Science and Technology, Malaysia, through research grants 6369101 and 6364002, respectively.
References
1. Alemnesh W, Hair-Bejo M, Aini I, Omar AR. Pathogenicity of fowl adenovirus in specific pathogen free chicken embryos. J Comp Pathol. 2012; 146:223–229.

2. Alvarado IR, Villegas P, El-Attrache J, Jensen E, Rosales G, Perozo F, Purvis LB. Genetic characterization, pathogenicity, and protection studies with an avian adenovirus isolate associated with inclusion body hepatitis. Avian Dis. 2007; 51:27–32.

3. Bancroft JD, Stevens A. Theory and Practice of Histological Techniques. 4th ed. Edinburgh: Churchill Livingstone;1996.
4. Benko M, Harrach B, Both GW, Russell WC, Adair BM, Adam E, de Jong JC, Hess M, Johnson M. Family Adenoviridae. In : Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. London: Academic Press;2005. p. 213–228.
5. Domanska-Blicharz K, Tomczyk G, Smietanka K, Kozaczynski W, Minta Z. Molecular characterization of fowl adenoviruses isolated from chickens with gizzard erosions. Poult Sci. 2011; 90:983–989.

6. Grgić H, Krell PJ, Nagy E. Comparison of fiber gene sequences of inclusion body hepatitis (IBH) and non-IBH strains of serotype 8 and 11 fowl adenoviruses. Virus Genes. 2014; 48:74–80.

7. Grgić H, Yang DH, Nagy E. Pathogenicity and complete genome sequence of fowl adenovirus serotype 8 isolate. Virus Res. 2011; 156:91–97.

8. Hair-Bejo M. Inclusion body hepatitis in a flock of commercial broiler chickens. J Vet Malaysia. 2005; 17:23–26.
9. Itakura C, Matsushita S, Goto M. Fine structure of inclusion bodies in hepatic cells of chickens naturally affected with inclusion body hepatitis. Avian Pathol. 1977; 6:19–32.

10. Juliana MA, Nurulfiza I, Hair-Bejo M, Omar AR, Aini I. Molecular characterization of fowl adenoviruses isolated from inclusion body hepatitis outbreaks in commercial broiler chickens in Malaysia. Pertanika J Trop Agric Sci. 2014; 37:483–497.
11. Kajan GL, Kecskeméti S, Harrach B, Benkő M. Molecular typing of fowl adenoviruses, isolated in Hungary recently, reveals high diversity. Vet Microbiol. 2013; 167:357–363.

12. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23:2947–2948.

13. Maartens LH, Joubert HW, Aitchison H, Venter EH. Inclusion body hepatitis associated with an outbreak of fowl adenovirus type 2 and type 8b in broiler flocks in South Africa. J S Afr Vet Assoc. 2014; 85:e1–e5.

14. Mansoor MK, Hussain I, Arshad M, Muhammad G. Preparation and evaluation of chicken embryo-adapted fowl adenovirus serotype 4 vaccine in broiler chickens. Trop Anim Health Prod. 2011; 43:331–338.

15. Meulemans G, Boschmans M, Berg TP, Decaesstecker M. Polymerase chain reaction combined with restriction enzyme analysis for detection and differentiation of fowl adenoviruses. Avian Pathol. 2001; 30:655–660.

16. Morshed R, Hosseini H, Langeroudi AG, Fard MHB, Charkhkar S. Fowl adenoviruses D and E cause inclusion body hepatitis outbreaks in broiler and broiler breeder pullet flocks. Avian Dis. 2017; 61:205–210.

17. Norina L, Norsharina A, Nurnadiah AH, Redzuan I, Ardy A, Nor-Ismaliza I. Avian adenovirus isolated from broiler affected with inclusion body hepatitis. Malaysian J Vet Res. 2016; 7:121–126.
18. Pallister J, Wright PJ, Sheppard M. A single gene encoding the fiber is responsible for variations in virulence in the fowl adenoviruses. J Virol. 1996; 70:5115–5122.

19. Raue R, Hess M. Hexon based PCRs combined with restriction enzyme analysis for rapid detection and differentiation of fowl adenoviruses and egg drop syndrome virus. J Virol Methods. 1998; 73:211–217.

20. Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938; 27:493–497.
21. Steer PA, O'Rourke D, Ghorashi SA, Noormohammadi AH. Application of high-resolution melting curve analysis for typing of fowl adenoviruses in field cases of inclusion body hepatitis. Aust Vet J. 2011; 89:184–192.

22. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28:2731–2739.

23. Wang Z, Wang B, Lou J, Yan J, Gao L, Geng R, Yu B. Mutation in fiber of adenovirus serotype 5 gene therapy vector decreases liver tropism. Int J Clin Exp Med. 2014; 7:4942–4950.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download