Abstract
Retinal pigment epithelium (RPE) is a major component of the eye. This highly specialized cell type facilitates maintenance of the visual system. Because RPE loss induces an irreversible visual impairment, RPE generation techniques have recently been investigated as a potential therapeutic approach to RPE degeneration. The microRNA-based technique is a new strategy for producing RPE cells from adult stem cell sources. Previously, we identified that antisense microRNA-410 (anti-miR-410) induces RPE differentiation from amniotic epithelial stem cells. In this study, we investigated RPE differentiation from umbilical cord blood-derived mesenchymal stem cells (UCB-MSCs) via anti-miR-410 treatment. We identified miR-410 as a RPE-relevant microRNA in UCB-MSCs from among 21 putative human RPE-depleted microRNAs. Inhibition of miR-410 induces overexpression of immature and mature RPE-specific factors, including MITF, LRAT, RPE65, Bestrophin, and EMMPRIN. The RPE-induced cells were able to phagocytize microbeads. Results of our microRNA-based strategy demonstrated proof-of-principle for RPE differentiation in UCB-MSCs by using anti-miR-410 treatment without the use of additional factors or exogenous transduction.
Defects in various neural retina subtypes result in eye disorders and loss of vision in many cases [17]. In terms of retinal degeneration, age-related macular degeneration (AMD) is a common cause in industrialized countries, and the advanced form of AMD can be divided into neovascular and non-neovascular atrophic types [1617]. In atrophic AMD, a gradual degeneration of retinal pigment epithelium (RPE) results in visual impairment. Cone–rod dystrophy is a type of retinal degeneration disease that causes a depletion of the outer nuclear layer, which consists of cone and rod photoreceptor cells. In addition to RPE cells, both types of photoreceptors are involved in visual signal transduction, as confirmed by their response to light. However, once photoreceptor or RPE cells are lost, these cell types are unable to regenerate themselves. Commonly used therapeutic applications for retinal degeneration retard the progression of degeneration by preventing neovascularization or through laser coagulation. However, as degenerated photoreceptor and RPE cells are unable to regenerate, the use of cell replacement therapy for vision restoration has recently been under investigation.
Many studies of RPE generation are based on embryonic stem (ES) or induced pluripotent stem (iPS) cells. It has even been suggested that pluripotent stem cells can spontaneously differentiate into RPE cells, with RPE cells spontaneously differentiating from ES or iPS cells in humans [1]. Moreover, it was recently reported that ES or iPS cells can differentiate into RPE-like cells following treatment with RPE development-relevant proteins [21420]. A relatively low yield of human RPE cells has been improved through the use of WNT and Nodal antagonists [13]; indeed, WNT signaling pathways are reported to antagonistically influence neural development [6811]. Furthermore, recent publications have demonstrated that nicotinamide and Activin A can enhance the RPE differentiation efficiency of human ES cells to achieve RPE differentiation within 4 weeks or in long-term cell culture [910]. Those RPE cells survived in a host eye and rescued retinal function after subretinal transplantation in a rat model of retinal degeneration. However, a clinical study using pluripotent stem cell-derived RPE cells suggested a severe risk to patients, such as teratoma formation of the remaining undifferentiated ES cells in the host after transplantation [22]. In studies to improve the safety of cell replacement therapy, such as sorting of undifferentiated cells by using SOX1 or SSEA-5 and long-term culturing for differentiation [2122], somatic stem cells have emerged as a useful source for clinical use.
Our previous study showed that miR-410 inhibition could induce RPE differentiation from human amniotic epithelial stem cells (AESCs) [5]. In addition, we showed that miR-410 is expressed at a relatively high level in human AESCs and that the inhibition of miR-410 induces mature RPE-specific factors. These cells were altered in morphology to form a cobblestone-like shape and they exhibited phagocytosis as is shown by ARPE-19 cells, a human RPE cell line that shows structural and functional RPE characteristics [7]. We further showed that miR-410 is predicted to directly target two RPE development-specific genes, RPE65 and OTX2. Our previous results indicated that a miRNA-based strategy can induce RPE differentiation from AESCs via treatment with a miR-410 inhibitor but without the use of additional factors and exogenous overexpression. Those results support a new hypothesis that microRNA inhibition might facilitate induction of RPE differentiation from other human somatic stem cells such as UCB-MSCs.
In this study, we suggest a new miR-based strategy to differentiate UCB-MSCs into functional RPE-like cells. We show that miR-410 is enriched in UCB-MSCs and that treatment with a miR-410 inhibitor can induce RPE differentiation by derepressing RPE65 and OTX2. Moreover, UCB-MSC-derived RPE-like cells were able to phagocytize, indicating their functionality as RPE cells. Our results show that miR-410 can serve as a useful tool to direct RPE differentiation from somatic stem cells by regulating multiple key genes that, as targets of miR, are suppressed.
Human UCB-MSCs and ARPE-19 cells were cultured as previously described [51518]. For RPE differentiation of human UCB-MSCs and AESCs, cells were cultured on culture dishes with the cell culture medium described above. After reaching 80% confluency, 100 ng/mL Activin A (R&D Systems, USA) and 10 mM nicotinamide (Sigma-Aldrich, USA) were supplemented to the cell culture medium. The medium was changed every 3 days for 9 weeks. For RPE differentiation by miRNAs, cells were transfected with microRNA or microRNA inhibitor as previously described [5]. Anti-miR microRNA inhibitor (AM11119; Ambion, USA) was designed to target has-miR-410-3p. For miR-induced RPE-like cells, human UCB-MSCs were transfected with anti-miR-410 four times (on days 0, 3, 10, and 17).
Total RNA was extracted from whole cell lysates and a human microRNA microarray (G4851a; Agilent Technologies, USA) procedure was performed. For data normalization and further analyses, GeneSpring GX software (ver. 11.5.1; Agilent Technologies) was used.
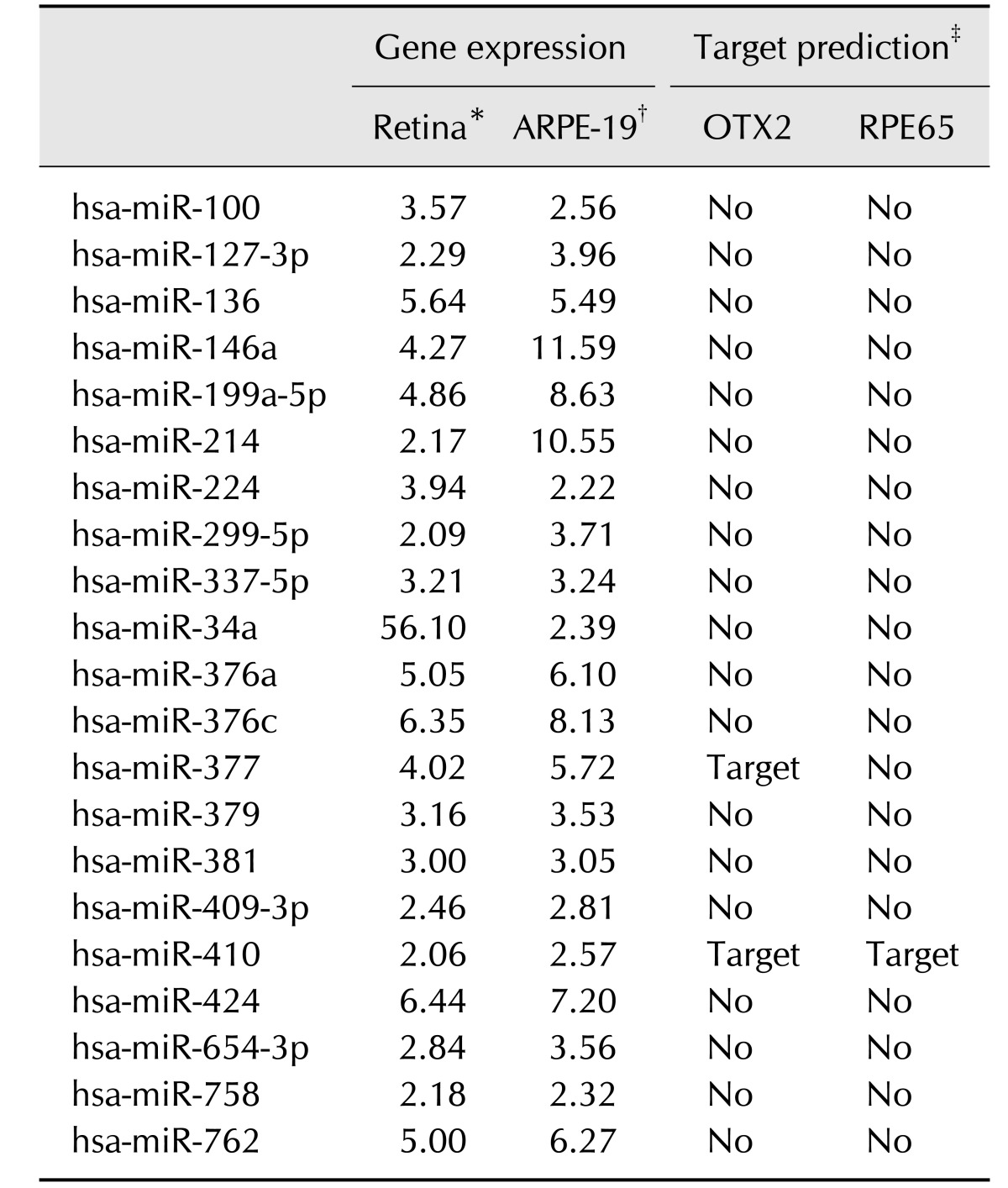
Using online microRNA target prediction programs, the predicted targets of hsa-miR-410-3p (5′-AAUAUAACACAG AUGGCCUGU-3′) were analyzed. To predict targets of microRNAs, we performed microRNA target prediction by using three different programs: TargetScan, miRanda, and DIANA. We specifically examined RPE development-relevant genes [12]: RPE-specific transcription factors (PAX6, MITF, OTX2), pigment synthesis-specific gene (TYRP1), membrane-associated proteins (BEST1, ZO-1, PEDF), visual cycle-specific genes (RPE65, LRAT, CRALBP), and phagocytosis-specific genes (MERTK, GULP1, LAMP2, VDP).
Total RNA was extracted from whole cell lysates. For quantitative analysis, gene expressions in each sample were normalized to the RPL13A gene and the relative expression level calculated by using the 2−ΔΔCt method. All of the primers used are shown in Supplementary Table 1.
For Western blot analysis, the antibodies used were: goat anti-mouse Alexa fluor 488 (Invitrogen, USA), GAPDH (Chemicon, USA), EMMPRIN (Abcam, UK) and Bestrophin (Millipore, USA). For counterstaining, nuclei were stained as blue with DAPI (Santa Cruz Biotechnology, USA).
After inducing RPE differentiation by using the microRNA transfections described above, cells were incubated with FluoSpheres carboxylate-modified microspheres (Invitrogen) as previously described [5]. For counterstaining, nuclei were stained as blue with DAPI (Sigma-Aldrich). For quantification, the number of bead-phagocytizing cells were counted.
Our previous study showed that miR-410 inhibition could induce RPE differentiation from AESCs [5]. We then hypothesized that microRNA inhibitions might facilitate an induction of RPE from UCB-MSCs. After completion of two microRNA microarrays with UCB-MSCs, human retina tissue and ARPE-19 cells, we identified the candidate microRNAs, which are enriched in UCB-MSCs and related to RPE-specific genes. The former microarray of UCB-MSCs versus human retina tissue revealed 51 microRNAs that had higher expression levels in UCB-MSCs than in retina (panel A in Fig. 1). In the same manner, we identified 28 microRNAs in the latter microarray analysis of UCB-MSCs versus ARPE-19 cells. Based on the results of these two microarray analyses, we selected 21 candidate microRNAs that were enriched in UCB-MSCs but were not enriched in either retina or ARPE-19 cells (panel B in Fig. 1).
To identify the most relevant microRNA, we performed further selections within the 21 candidate microRNAs by using three different microRNA target prediction programs in order to identify microRNAs that target more than five genes in the RPE development process and that are predicted to target two known RPE-specific factors, RPE65 and OTX2 (Table 1). Interestingly, as in our AESC study [5], miR-410 was the strongest candidate in UCB-MSCs and, thus, was chosen for further study. To confirm the microarray results, we compared expression levels of miR-410 among UCB-MSCs, anti-miR-410-treated UCB-MSCs, and ARPE-19 cells (panel C in Fig. 1). The expression level of miR-410 in the anti-miR-410-treated UCB-MSCs was significantly reduced from the level in the control microRNA-treated UCB-MSCs. Thus, we used anti-miR-410 to derepress RPE-specific genes in terms of induction of a direct differentiation of UCB-MSCs into RPE-like cells.
To induce RPE differentiation in UCB-MSCs, we cultured cells with anti-miR-410 for 3 weeks or in RPE differentiation medium including 100 ng/mL Activin A and 10 mM nicotinamide for 9 weeks (panel A in Fig. 2). To characterize the RPE-induced cells, conventional and quantitative gene expression analyses were performed by using total RNA from the two-factor (2F)-induced UCB-MSCs after 9 weeks and from the anti-miR-410-treated UCB-MSCs after 2 and 21 days. Transcripts of the RPE progenitor factor MITF and four mature RPE-relevant factors RPE65, LRAT, Bestrophin, and EMMPRIN were increased in 2F-induced RPE-like cells (panel B in Fig. 2). After RPE differentiation with anti-miR-410 transfections, the levels of all transcripts were significantly increased over that of the controls. The anti-miR-410-treated UCB-MSCs after the first transfection showed increased MITF, Bestrophin, and EMMPRIN gene expression levels. These results indicate that consecutive treatments of anti-miR-410 can induce direct RPE differentiation from UCB-MSCs.
We then analyzed protein expressions in the RPE-like cells derived from human UCB-MSCs. First, western blot analyses of RPE-specific factors in the 2F-induced RPE-like cells and the anti-miR-410-induced RPE-like cells were performed. From both sources of RPE-like cells, we detected the mature RPE-specific factors Bestrophin and EMMPRIN (panel C in Fig. 2). A homolog of western blot analysis, immunocytochemistry results revealed significantly increased numbers of Bestrophin- and EMMPRIN-positive cells as a result of 2F or anti-miR-410 treatments in UCB-MSCs (panels A-C in Fig. 3). To determine the functional ability of the RPE-like cells from UCB-MSCs, a phagocytosis assay was performed after anti-miR-410 treatment (panel A in Fig. 4). As previously demonstrated [7], ARPE-19 cells showed a high level of phagocytosis, which was indicated by the internalizing of most of the beads (panel B in Fig. 4). In this analysis of phagocytic ability, the number of bead-internalizing cells among the UCB-MSCs was significantly increased upon anti-miR-410 treatment than in those without treatment. These results demonstrate that miR-induced RPE-like cells from UCB-MSCs exhibit similar levels of gene expressions and phagocytosis to those of ARPE-19 cells.
In this study, we investigated a technique to induce RPE differentiation of UCB-MSCs by using miR-410 inhibition. Expression of miR-410 is relatively high in UCB-MSCs and is higher than that in retina tissue or RPE; moreover, miR-410 targets multiple RPE-specific genes. The miR-410 inhibition was able to increase the gene and protein expressions of RPE-specific factors and to induce phagocytic capabilities in UCB-MSCs. Consequently, miR-410 inhibition seems to be able to induce RPE differentiation. Our results indicate the potential for direct differentiation of UCB-MSCs into RPE-like cells by using miR-410 inhibition.
As reported in our previous study, miR-410 can directly target two RPE development-relevant factors: RPE65 and OTX2 [5]. In that study, we demonstrated that anti-miR-410 significantly reduced the endogenous level of miR-410 in AESCs and that it can decrease both gene and protein expression levels of OTX2 and RPE65 by binding to complementary sites in the 3′UTR. Moreover, the decreased expression of genes could be derepressed by the microRNA-mediated inhibition, anti-miR-410.
Apart from the RPE differentiation, other roles of miR-410 have been described in several recent studies. Two studies demonstrated that miR-410 in tumor cells has a tumor-suppressive role by targeting MET or CDK1 [3 4]. Chien et al. reported that the expression level of CDK1 is regulated upon overexpression or inhibition of miR-410/-650 and that the expression of both miRNAs can be induced by p16INK4a overexpression. Those authors concluded that tumor suppressor p16INK4a inhibits the expression of CDK1 through miR-410/-650 in the MCF7 carcinoma and U87 glioma cell lines. Chen et al. [3] reported that miR-410 directly targets MET, a hepatocyte growth factor receptor, and subsequently regulates the proliferation and invasion of human glioma. In another cell type, miR-410 was found to have a role as a regulator of tissue regeneration. Snyder et al. [19] demonstrated that overexpressions of miR-410 and miR-433 induce myogenic differentiation in Mef2a-deficient myoblasts, with the expression level of secreted frizzled-related proteins (sFRPs) being increased, whereas WNT activity is decreased. These two miRNAs directly target and simultaneously repress sFRP2, a WNT signaling inhibitor. The authors commented that miR-mediated regulation of WNT signaling could have potential use in muscle regeneration.
Together with our previous study of RPE differentiation from AESCs, the present proof-of-principle experiment is the first demonstration of RPE direct differentiation in somatic stem cells through the effects of a single microRNA. Our results indicate that miR-410 inhibition directly regulates two RPE-specific factors, OTX2 and RPE65, and induces RPE-specific gene and protein expressions in somatic stem cells. The results of this study suggest that miR-410 inhibition can regulate multiple RPE-relevant genes and have a useful role in direct RPE differentiation from human UCB-MSCs. In the near future, this microRNA-based technique may serve as an attractive strategy for curing human retinal degenerative diseases.
Acknowledgments
This research was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant No. HI14C3301), and partially supported by the Research Institute for Veterinary Science, Seoul National University (Republic of Korea).
References
1. Aoki H, Hara A, Nakagawa S, Motohashi T, Hirano M, Takahashi Y, Kunisada T. Embryonic stem cells that differentiate into RPE cell precursors in vitro develop into RPE cell monolayers in vivo. Exp Eye Res. 2006; 82:265–274. PMID: 16150443.

2. Buchholz DE, Hikita ST, Rowland TJ, Friedrich AM, Hinman CR, Johnson LV, Clegg DO. Derivation of functional retinal pigmented epithelium from induced pluripotent stem cells. Stem Cells. 2009; 27:2427–2434. PMID: 19658190.

3. Chen L, Zhang J, Feng Y, Li R, Sun X, Du W, Piao X, Wang H, Yang D, Sun Y, Li X, Jiang T, Kang C, Li Y, Jiang C. MiR-410 regulates MET to influence the proliferation and invasion of glioma. Int J Biochem Cell Biol. 2012; 44:1711–1717. PMID: 22750473.

4. Chien WW, Domenech C, Catallo R, Kaddar T, Magaud JP, Salles G, Ffrench M. Cyclin-dependent kinase 1 expression is inhibited by p16INK4a at the post-transcriptional level through the microRNA pathway. Oncogene. 2011; 30:1880–1891. PMID: 21170085.
5. Choi SW, Kim JJ, Seo MS, Park SB, Kang TW, Lee JY, Lee BC, Kang I, Shin TH, Kim HS, Yu KR, Kang KS. miR-410 inhibition induces RPE differentiation of amniotic epithelial stem cells via overexpression of OTX2 and RPE65. Stem Cell Rev. 2015; 11:376–386. PMID: 25351180.

6. Diep DB, Hoen N, Backman M, Machon O, Krauss S. Characterisation of the Wnt antagonists and their response to conditionally activated Wnt signalling in the developing mouse forebrain. Brain Res Dev Brain Res. 2004; 153:261–270. PMID: 15527894.

7. Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res. 1996; 62:155–169. PMID: 8698076.

8. Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998; 391:357–362. PMID: 9450748.

9. Idelson M, Alper R, Obolensky A, Ben-Shushan E, Hemo I, Yachimovich-Cohen N, Khaner H, Smith Y, Wiser O, Gropp M, Cohen MA, Even-Ram S, Berman-Zaken Y, Matzrafi L, Rechavi G, Banin E, Reubinoff B. Directed differentiation of human embryonic stem cells into functional retinal pigment epithelium cells. Cell Stem Cell. 2009; 5:396–408. PMID: 19796620.

10. Kokkinaki M, Sahibzada N, Golestaneh N. Human induced pluripotent stem-derived retinal pigment epithelium (RPE) cells exhibit ion transport, membrane potential, polarized vascular endothelial growth factor secretion, and gene expression pattern similar to native RPE. Stem Cells. 2011; 29:825–835. PMID: 21480547.

11. Lamb TM, Knecht AK, Smith WC, Stachel SE, Economides AN, Stahl N, Yancopolous GD, Harland RM. Neural induction by the secreted polypeptide noggin. Science. 1993; 262:713–718. PMID: 8235591.

12. Liao JL, Yu J, Huang K, Hu J, Diemer T, Ma Z, Dvash T, Yang XJ, Travis GH, Williams DS, Bok D, Fan G. Molecular signature of primary retinal pigment epithelium and stem-cell-derived RPE cells. Hum Mol Genet. 2010; 19:4229–4238. PMID: 20709808.

13. Osakada F, Ikeda H, Mandai M, Wataya T, Watanabe K, Yoshimura N, Akaike A, Sasai Y, Takahashi M. Toward the generation of rod and cone photoreceptors from mouse, monkey and human embryonic stem cells. Nat Biotechnol. 2008; 26:215–224. PMID: 18246062.

14. Osakada F, Ikeda H, Sasai Y, Takahashi M. Stepwise differentiation of pluripotent stem cells into retinal cells. Nat Protoc. 2009; 4:811–824. PMID: 19444239.

15. Park SB, Yu KR, Jung JW, Lee SR, Roh KH, Seo MS, Park JR, Kang SK, Lee YS, Kang KS. bFGF enhances the IGFs-mediated pluripotent and differentiation potentials in multipotent stem cells. Growth Factors. 2009; 27:425–437. PMID: 19919531.

16. Pascolini D, Mariotti SP. Global estimates of visual impairment: 2010. Br J Ophthalmol. 2012; 96:614–618. PMID: 22133988.

17. Resnikoff S, Pascolini D, Etya'ale D, Kocur I, Pararajasegaram R, Pokharel GP, Mariotti SP. Global data on visual impairment in the year 2002. Bull World Health Organ. 2004; 82:844–851. PMID: 15640920.
18. Roh DH, Seo MS, Choi HS, Park SB, Han HJ, Beitz AJ, Kang KS, Lee JH. Transplantation of human umbilical cord blood or amniotic epithelial stem cells alleviates mechanical allodynia after spinal cord injury in rats. Cell Transplant. 2013; 22:1577–1590. PMID: 23294734.

19. Snyder CM, Rice AL, Estrella NL, Held A, Kandarian SC, Naya FJ. MEF2A regulates the Gtl2-Dio3 microRNA mega-cluster to modulate WNT signaling in skeletal muscle regeneration. Development. 2013; 140:31–42. PMID: 23154418.
20. Sonoda S, Spee C, Barron E, Ryan SJ, Kannan R, Hinton DR. A protocol for the culture and differentiation of highly polarized human retinal pigment epithelial cells. Nat Protoc. 2009; 4:662–673. PMID: 19373231.

21. Tang C, Lee AS, Volkmer JP, Sahoo D, Nag D, Mosley AR, Inlay MA, Ardehali R, Chavez SL, Pera RR, Behr B, Wu JC, Weissman IL, Drukker M. An antibody against SSEA-5 glycan on human pluripotent stem cells enables removal of teratoma-forming cells. Nat Biotechnol. 2011; 29:829–834. PMID: 21841799.

22. Wolber W, Ahmad R, Choi SW, Eckardt S, McLaughlin KJ, Schmitt J, Geis C, Heckmann M, Sirén AL, Müller AM. Phenotype and stability of neural differentiation of androgenetic murine ES cell-derived neural progenitor cells. Cell Med. 2013; 5:29–42. PMID: 26858862.

Fig. 1
Putative retinal pigment epithelium (RPE)-specific microRNAs in human umbilical cord blood-derived mesenchymal stem cells (UCB-MSCs). (A) Microarray data indicate the number of microRNAs that are expressed at a higher level in UCB-MSCs than in retina tissue or ARPE-19 cells. Of these, 21 microRNAs belong to both groups. (B) Number of predicted targets for the 21 putative microRNAs determined by using three different target prediction programs; TargetScan, miRanda, and DIANA. (C) Results of real-time RT-PCR analysis of miR-410 expression in human UCB-MSCs (miR ctl), anti-miR-410-treated cells, and the human RPE cell line ARPE-19. ***p < 0.001.
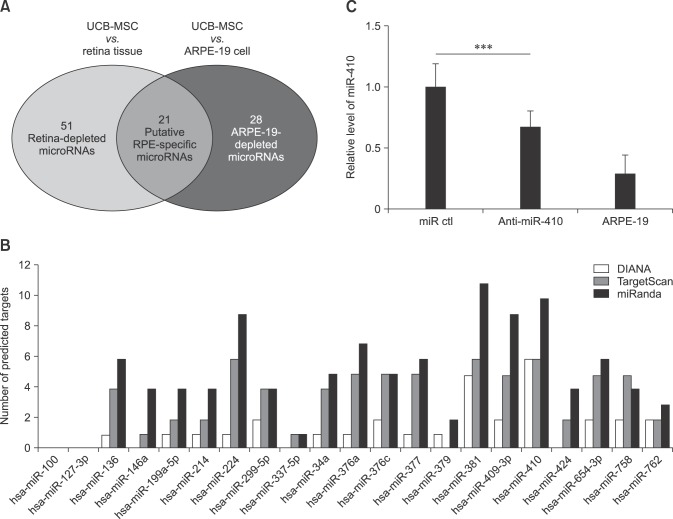
Fig. 2
Induction of RPE differentiation from human umbilical cord blood-derived mesenchymal stem cells (UCB-MSCs) by miR-410 inhibition. (A) Schematic of the differentiation protocol for UCB-MSCs into retinal pigment epithelium (RPE)-like cells via 2F induction for 9 weeks or by microRNA transfections at days 0, 3, 10 and 17. (B) Conventional RT-PCR measurement of the expression of RPE-specific factors in UCB-MSCs post-induction with 2F and post-treatment with miR control or anti-miR-410 at days 2 and 21. (C) Western blot results for RPE-specific factors Bestrophin and EMMPRIN performed for 2F-induced RPE-like cells and miR-induced RPE-like cells.
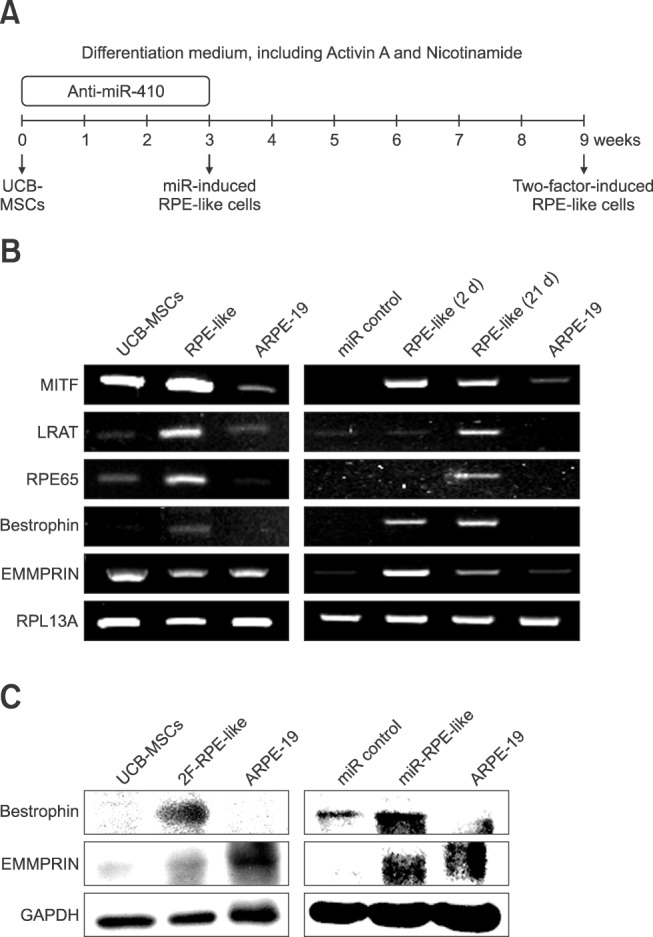
Fig. 3
Retinal pigment epithelium (RPE)-specific protein expression in RPE-like cells. (A and B) Representative immunocytochemistry images showing that 2F-induced and anti-miR-410-treated RPE-like cells expressed the mature RPE markers Bestrophin and EMMPRIN (both green). Nuclei were counterstained with DAPI. (C) Quantification of Bestrophin- and EMMPRIN-positive cells in miR control, anti-miR-410-treated UCB-MSCs, and ARPE-19 cells. *p < 0.05, ***p < 0.001. Scale bar = 0.2 mm (A and B).
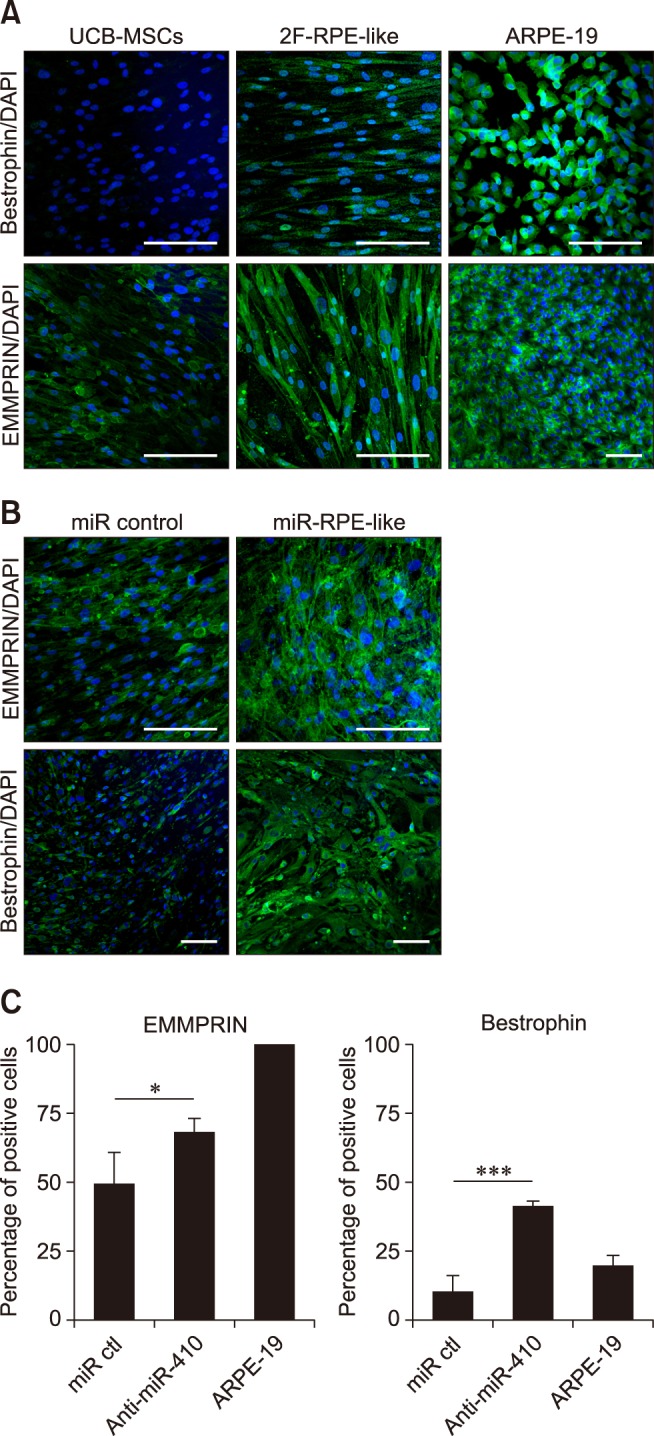
Fig. 4
Enhanced phagocytosis of anti-miR-410-treated umbilical cord blood-derived mesenchymal stem cells (UCB-MSCs). (A) Representative images of phagocytosis assay results using fluorescent microsphere beads 21 days after transfection of miR control or anti-miR-410 into UCB-MSCs. Treatments of anti-miR-410 in human UCB-MSCs increased the internalization of red fluorescent beads. (B) Quantification of bead phagocytosis level after treatments of anti-miR-410 for 21 days. ***p < 0.001. Scale bars = 50 µm (upper panels), 5 µm (lower panels).




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download