Abstract
Translocations involving chromosome 21q22 are frequently observed in hematologic malignancies including acute myeloid leukemia (AML), most of which have been known to be involved in malignant transformation through transcriptional dysregulation of Runt-related transcription factor 1 (RUNX1) target genes. Nineteen RUNX1 translocational partner genes, at least, have been identified, but not Homeobox A (HOXA) genes so far. We report a novel translocation of RUNX1 into the HOXA gene cluster in a 57-year-old female AML patient who had been diagnosed with myelofibrosis 39 months ahead. G-banding showed 46,XX,t(7;21)(p15;q22). The involvement of RUNX1 and HOXA genes was confirmed by fluorescence in situ hybridization.
Translocation of chromosome 21q22 resulting in alterations of the Runt-related transcription factor 1 (RUNX1) gene commonly occurs in hematologic malignancies. The RUNX1 gene (known as AML-1, CBFA2, or PEBP2αB) encodes the α-subunit of core binding factor (CBFα) which forms heterodimer with core-binding factor-β (CBFβ) and plays a key role in the initiation of hematopoiesis and the generation of hematopoietic stem cells in the embryo (1,2). The RUNX1 gene was first identified as a fusion partner in the t(8;21) (q22;q22) in AML-M2 (3). To date, 39 sites of recurrent translocation have been described in hematologic malignancies, and a minimum of 19 partner genes have been identified at the molecular level (4).
HOX genes were first identified as essential factors for the regulation of limb positioning during embryogenesis (5). The HOX gene family, which encodes DNA-binding transcription factors, consists of 39 HOX genes arranged into 4 gene clusters (A, B, C, and D) located on 4 different chromosomes, 7p15, 17q21, 12q13 and 2q31, respectively. HOX A, B, and C clustered genes are expressed during early hematopoiesis of hematopoietic stem cells and progenitors, and are downregulated during the later stages of differentiation and maturation of hematopoietic progenitors (6). It has been reported that dysregulation of HOX genes could have an impact on leukemic development (7,8). The NUP98-HOXA9 fusion protein has been demonstrated to play a role in leukemogenesis (9), which indicates that translocation and fusion protein generation could be a mechanism responsible for altered HOX gene function.
The current case, to the best of our knowledge, is the first report demonstrating juxtaposition of HOXA and RUNX via reciprocal translocation, t(7;21)(p15;q22), in AML.
A sample of bone marrow aspirate from a 57-year-old woman with a presumptive diagnosis of AML was submitted for karyotype analysis and fluorescence in situ hybridization (FISH). She had been diagnosed with chronic idiopathic myelofibrosis 39 months prior to this presentation. The complete blood count (CBC) indicated the following: Hb 75 g/L; platelet count 41×109/L; and leukocyte count 50.6×109/L with 10% neutrophils, 59% lymphocytes, 2% metamyelocytes, and 29% blasts. Bone marrow aspirate smears showed 100% cellularity with 67% blasts which consisted of moderate-sized to-large cells showing high nuclear/cytoplasmic ratios, fine nuclear chromatin, and prominent nucleoli, but no definite Auer rods. Immunophenotype analysis revealed that leukemic cells were strongly positive for HLA-DR, CD34, CD13, CD33, CD117, and myeloperoxidase.
Conventional cytogenetic analysis was performed in bone marrow cells by using unstimulated 24-h culture method and G-banding. The karyotype was described according to the guidelines of International System for Human Cytogenetic Nomenclature (ISCN) 2009.
To evaluate the status of RUNX1 on 21q22, FISH was performed with a commercial LSI AML1/ETO Dual Color, Dual Fusion Translocation Probe (Vysis, Downers Grove, IL). To investigate the breakpoint on the short arm of chromosome 7 inducing the t(7;21), FISH was performed with "in-house" dual color break-apart BAC probes designed to target the specific sites of HOXA-clustered genes on chromosome 7p15. Eight BAC clones specific for the HOXA genes were selected from the human genome browser database Genome Bioinformatics Group, University of California, Santa Cruz (http://genome.ucsc.edu/) (Fig. 2A); 3 clones (RP11-838G2, RP11-1080K8 and RP11-627P22) and 2 clones (RP11-1148E13 and RP11-299F5) targeting the telomeric and centromeric side of the HOXA gene cluster, respectively, and the remaining 3 clones (RP11-163M21, RP11-1132K14, and RP11-1025G19) alternatively encompassing the HOXA genes. After bacterial culture, plasmid DNA was purified using the plasmid DNA purification kit (Qiagen Sciences, Germantown, MD), and labeled by nick-translation (Nick Translation Kit; Abbott Molecular Inc., North Chicago, IL). FISH with "in house" BAC probes was performed as previously reported (10). Briefly, probe signals were enumerated from 200 interphase cells. A cutoff for a true positive break apart signal was set at >5% of interphase cells showing separation of red and green signals at a level greater than twice the usual signals. All probes were verified for the expected site of hybridization by using normal lymphocyte metaphases (data not shown).
Interphase FISH using the LSI AML1/ETO probes showed 1 large and 2 small green signals, and 2 red signals in ~50% of the nuclei. Metaphase FISH analysis demonstrated 1 large green signal on the intact chromosome 21 and 2 small green signals on the derivative chromosome 7 as well as on derivative chromosome 21 (Fig. 1B). These results indicate that 1 RUNX1 gene split and translocated to a position other than the ETO gene, which is consistent with the results of cytogenetic analysis, that is, t(7;21)(p15;q22).
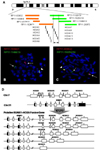
Five pairs of break-apart probe sets targeting the HOXA gene cluster were applied to normal control slides and patient bone marrow slides. Every probe set showed fusion or closely paired signals in normal lymphocytes, whereas the patient slides revealed fusion or separated signals depending on the probe set combinations. FISH using probe set 1 (RP11-838G2, RP11-1148E13), probe set 2 (RP11-1080K8, RP11-299F5), and probe set 3 (RP11-163M21, RP11-1148E13) demonstrated 1 closely paired red-green fusion signal on the normal chromosome 7, but 1 red signal on the derivative chromosome 21 and 1 green signal on the derivative chromosome 7 in the metaphase cells (Fig. 2B). These observations indicate that the break point is within the HOXA gene. One fusion, 1 red, and 1 green signals were found in ~35% of the interphase cells. FISH using probe set 4 (RP11-838G2, RP11-1025G19) and probe set 5 (RP11-627P22, RP11-1132K14) showed 1 fusion signal each on the normal chromosome 7 and the derivative chromosome 21, and 1 small green signal on the derivative chromosome 7 in the patient metaphase cells (Fig. 2C). These results indicate that the break point is located in the region covered by both RP11-1025G19 and RP11-1132K14 probes. Interphase FISH showed the same signal pattern in 30% of the bone marrow cells. Taken together, these results narrow down the break point to within 50 kb of the region comprising HOXA 9-13 genes (Fig. 2A).
A novel translocation of RUNX1 to the HOXA gene cluster in an AML patient with 46,XX,t(7;21)(p15;q22) was identified, which implies that HOXA and RUNX1 could be reciprocal translocational gene partners.
These cases of AML with t(7;21)(p15;q22) has been reported: 1 case with a single translocation and the other 2 cases with complex chromosomal aberrations (11,12). More than 30 cancer-associated candidate genes, including the HOXA gene cluster, have been identified on chromosome 7p15 (4). However, the identity of the gene on the 7p15 section of the chromosome that is the translocation partner of RUNX1 remains unclear.
HOXA cluster genes play an important role in homeostatic hematopoiesis, and their deregulation can induce leukemic transformation (6-8). Several mechanisms have been suggested for HOX gene deregulation as a single or co-operative factor in leukemogenic function (8,13). NUP98-HOXA9 gene expression is leukemogenic in mice (7) and is detected in human AML cells (9,14). These reports show that translocation and fusion protein generation could be a mechanism underlying HOX gene deregulation. No reports of a translocational partnership between the HOXA gene cluster and RUNX1 have been published to date.
The current data show a definite reciprocal translocation between RUNX1 and the HOXA gene cluster. FISH data with a BAC clone probe set combination localized the gene fusion site to a 50-kb region, which comprises HOXA9, HOXA10, HOXA11, and HOXA13 (Fig. 2A). Considering frequently occurring break points within both genes as well as the current data, possible translocational fusion genes, including RUNX1-HOXA9, RUNX1-HOXA10, RUNX1-HOXA11 , and RUNX1-HOXA13, can be postulated (Fig. 2D).
Efficient functioning of the transcription factor CBFα, encoded by RUNX1, normally requires its heterodimeric partner CBFβ which promotes DNA binding of CBFα (15). AML1-ETO, the prototype RUNX1 fusion gene product, functions as a dominant-negative transcription factor on CBFα target genes (16). NUP98-HOXA9 fusion protein binds to DNA via the homeodomain of HOXA9 and is thought to function as a transcription factor (8,9,14). The case under study did not show any other translocation except t(7;21)(p15;q22)/RUNX1-HOXA gene cluster, and all possible fusion genes HOXA(9-13) may be equally responsible for homeodomain formation. Taken together, these observations suggest that t(7;21)(p15;q22)/RUNX1-HOXA(9-13) may play a causative role in leukemic development in the patient under study.
Studies with animal models have demonstrated that artificially induced overexpression of HOX genes, including HOXA9, HOXA10, and NUP98-HOXA9, gave rise to myeloproliferative diseases, which then progressed to AML with long-latency periods (7,17). Secondary AML from myelofibrosis was fatal except 2 cases showing favorable cytogenetic abnormalities, t(8;21) and inv(16) (18). However, the exact role of the involved genes remains unclear. The present case was secondary AML from chronic myelofibrosis at an interval of 39 months. Unfortunately, it is unclear whether t(7;21) (p15;q22)/RUNX1-HOXA in the present case was associated with the early developmental stage of myeloproliferative neoplasms or with the late stage progression to secondary AML because the initial bone marrow sample was inadequate for cytogenetic studies, similar to most myelofibrotic bone marrow samples. Further identification of fusion gene products or direct sequencing of the fusion gene was not performed because of limited bone marrow samples.
In summary, this study described a rare translocation of t(7;21)(p15;q22) and the first identification of RUNX1-HOXA fusion gene involvement in AML, diagnosed secondary to myelofibrosis, which suggests the possibility that RUNX1 and HOXA genes could be reciprocal translocation partners and may play a role in leukemogenesis.
Figures and Tables
Figure 1
(A) Representative metaphase G-banding of the patient bone marrow aspirate showing 46,XX, t(7;21)(p22;q22). (B) Metaphase fluorescence in situ hybridization analysis using AML1/ETO dual-color, dual-fusion translocation probes demonstrate translocation of RUNX1 into chromosome 7; 2 usual-sized red signals on 2 intact chromosome 8, 1 usual-sized green signal on chromosome 21 (arrow), and tw1 small green signal each on derivative chromosome 21 (arrowhead) and derivative chromosome 7 (dotted arrow).
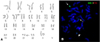
Figure 2
(A) BAC clone probes designed and HOXA gene clusters on chromosome 7p15.2. The arrow between the dotted and solid lines represents the break point area. (B) Metaphase FISH using the probe set RP11-163M21 and RP11-1148E13 showing translocation of the gene segment, including HOXA1-7, to derivative chromosome 21. (C) Metaphase FISH using probe set RP11-838G2 and RP11-1025G19 showing translocation of the gene segment, including a part of HOXA 9-13, to derivative chromosome 21. (D) Putative fusion gene map of t(7;21): RUNX1/HOXA9-13.
ACKNOWLEDGEMENTS
This work was supported by a Terry Fox Program Project Award, the Leukemia Lymphoma Society of Canada and the British Columbia Cancer Foundation.
References
1. Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996; 84:321–330.

2. de Bruijn MF, Speck NA. Core-binding factors in hematopoiesis and immune function. Oncogene. 2004; 23:4238–4248.

3. Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci USA. 1991; 88:10431–10434.

4. Huret JL. Complex translocations, simple variant translocations and Ph-negative cases in chronic myelogenous leukaemia. Hum Genet. 1990; 85:565–568.

6. Pineault N, Helgason CD, Lawrence HJ, Humphries RK. RK Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp Hematol. 2002; 30:49–57.

7. Kroon E, Thorsteinsdottir U, Mayotte N, Nakamura T, Sauvageau G. NUP98-HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. EMBO J. 2001; 20:350–361.

8. Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007; 26:6766–6776.

9. Chou WC, Chen CY, Hou HA, Lin LI, Tang JL, Yao M, Tsay W, Ko BS, Wu SJ, Huang SY, Hsu SC, Chen YC, Huang YN, Tseng MH, Huang CF, Tien HF. Acute myeloid leukemia bearing t(7;11)(p15;p15) is a distinct cytogenetic entity with poor outcome and a distinct mutation profile: comparative analysis of 493 adult patients. Leukemia. 2009; 23:1303–1310.

10. Makretsov N, He M, Hayes M, Chia S, Horsman DE, Sorensen PH, Huntsman DG. A fluorescence in situ hybridization study of ETV6-NTRK3 fusion gene in secretory breast carcinoma. Genes Chromosomes Cancer. 2004; 40:152–157.

11. Koo SH, Kwon GC, Chun HJ, Park JW. Cytogenetic and fluorescence in situ hybridization analyses of hematologic malignancies in Korea. Cancer Genet Cytogenet. 1998; 101:1–6.

12. Jeandidier E, Dastugue N, Mugneret F, Lafage-Pochitaloff M, Mozziconacci MJ, Herens C, Michaux L, Verellen-Dumoulin C, Talmant P, Cornillet-Lefebvre P, Luquet I, Charrin C, Barin C, Collonge-Rame MA, Perot C, Van den Akker J, Gregoire MJ, Jonveaux P, Baranger L, Eclache-Saudreau V, Pages MP, Cabrol C, Terre C, Berger R. Groupe Francais de Cytogenetique Hematologique. Abnormalities of the long arm of chromosome 21 in 107 patients with hematopoietic disorders: a collaborative retrospective study of the Groupe Francais de Cytogenetique Hematologique. Cancer Genet Cytogenet. 2006; 166:1–11.

13. Pineault N, Abramovich C, Ohta H, Humphries RK. Differential and common leukemogenic potentials of multiple NUP98-Hox fusion proteins alone or with Meis1. Mol Cell Biol. 2004; 24:1907–1917.

14. Yassin ER, Sarma NJ, Abdul-Nabi AM, Dombrowski J, Han Y, Takeda A, Yaseen NR. Dissection of the transformation of primary human hematopoietic cells by the oncogene NUP98-HOXA9. PLoS One. 2009; 4:e6719.

15. Wang Q, Stacy T, Miller JD, Lewis AF, Gu TL, Huang X, Bushweller JH, Bories JC, Alt FW, Ryan G, Liu PP, Wynshaw-Boris A, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996; 87:697–708.

16. Meyers S, Lenny N, Hiebert SW. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol Cell Biol. 1995; 15:1974–1982.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download