Abstract
Purpose
To investigate the methylation status of cancer-associated genes in breast cancer to assess its use in the diagnosis of breast cancer and the relationship with distinctive clinical and pathological features.
Methods
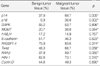
A total of 29 benign tumors and their adjacent normal tissues as well as 67 malignant tumors and adjacent normal samples, from women undergoing surgery for primary invasive breast carcinoma at Uijongbu St. Mary's Hospital, between March 2003 and March 2005, were used. Eleven candidate genes were chosen; P14, P16, DAPK, MGMT, h-MLH, E-cadherin, RASSF1α, Twist, RARβ, HIN-1, and Cyclin D. DNA was extracted from fresh tissues, and methylation specific PCR performed.
Results
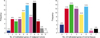
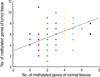
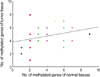
The number of methylated genes was increased in the malignant tissues compared to the benign tumors and adjacent normal tissues. 7 genes; P14, P16, MGMT, RASSF1α, Twist, RARβ, and Cyclin D, were more frequently methylated in malignant than benign tumors, with the differences in the p14, p16, and RARβ, genes were statistically significant (p<0.05). In benign tumors, the p16 and HIN-1 genes were the most infrequently (6.9%) and frequently methylated (82.8%), respectively. In malignant tumors, the h-MLH and RASSF1α genes were most infrequently and frequently methylated genes, respectively. The subgroup showing methylation of the DAPK gene had a higher nuclear grade and greater progesterone receptor negativity. The group in which the RASSF1α gene was methylated, had greater estrogen receptor (ER) and progesterone receptor (PgR) positivities. The Twist gene was frequently methylated in the subgroup showing higher nuclear and histologic grades. The group with HIN-1 and cyclin D methylation had a tendency to show greater ER positivity.
Conclusion
The subgroups showing methylated DAPK and Twist should be more intensely treated and followed up more carefully than those with RASSF1α, HIN-1 and Cyclin D methylation. Gene methylation may be linked to various pathological features of breast cancer; however, this will require confirmation from larger studies.
Figures and Tables
References
2. Garinis GA, Patrinos GP, Spanakis NE, Menounos PG. DNA hypermethylation: when tumour suppressor genes go silent. Hum Genet. 2002. 111:115–127.

3. Szyf M, Pakneshan P, Rabbani SA. DNA methylation and breast cancer. Biochem Pharmacol. 2004. 68:1187–1197.

4. Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene. 2002. 21:5462–5482.

5. Esteller M. Dormant hypermethylated tumour suppressor genes: questions and answers. J Pathol. 2005. 205:172–180.

6. van Rijnsoever M, Grieu F, Elsaleh H, Joseph D, Iacopetta B. Characterisation of colorectal cancers showing hypermethylation at multiple CpG islands. Gut. 2002. 51:797–802.

7. Yan PS, Perry MR, Laux DE, Asare AL, Caldwell CW, Huang TH. CpG island arrays: an application toward deciphering epigenetic signatures of breast cancer. Clin Cancer Res. 2000. 6:1432–1438.
8. Fackler MJ, McVeigh M, Evron E, Garrett E, Mehrotra J, Polyak K, et al. DNA methylation of RASSF1α, HIN-1, RAR-beta, Cyclin D2 and Twist in in situ and invasive lobular breast carcinoma. Int J Cancer. 2003. 107:970–975.

9. Lehmann U, Langer F, Feist H, Glockner S, Hasemeier B, Kreipe H. Quantitative assessment of promoter hypermethylation during breast cancer development. Am J Pathol. 2002. 160:605–612.

10. Garcia JM, Silva J, Pena C, Garcia V, Rodriguez R, Cruz MA, et al. Promoter methylation of the PTEN gene is a common molecular change in breast cancer. Genes Chromosomes Cancer. 2004. 41:117–124.
11. Bae YK, Brown A, Garrett E, Bornman D, Fackler MJ, Sukumar S, et al. Hypermethylation in histologically distinct classes of breast cancer. Clin Cancer Res. 2004. 10:5998–6005.

12. Mason SL, Loughran O. La Thangue NB. P14(ARF) regulates E2F activity. Oncogene. 2002. 21:4220–4230.
13. Enders GH, Koh J, Missero C, Rustgi AK, Harlow E. P16 inhibition of transformed and primary squamous epithelial cells. Oncogene. 1996. 12:1239–1245.
14. Liu Y, Lee MO, Wang HG, Li Y, Hashimoto Y, Klaus M, et al. Retinoic acid receptor beta mediates the growth-inhibitory effect of retinoic acid by promoting apoptosis in human breast cancer cells. Mol Cell Biol. 1996. 16:1138–1149.

15. Sabharwal A, Middleton MR. Exploiting the role of O(6)-methylguanine-DNA-methyltransferase (MGMT) in cancer therapy. Curr Opin Pharmacol. 2006. 6:355–363.

16. Burbee DG, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, Gao B, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001. 93:691–699.

17. Shapiro GI. Cyclin-dependent kinase pathways as targets for cancer treatment. J Clin Oncol. 2006. 24:1770–1783.

18. Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004. 117:927–939.

19. Cohen O, Feinstein E, Kimchi A. DAP-kinase is a Ca2+/calmodulin-dependent, cytoskeletal-associated protein kinase, with cell death-inducing functions that depend on its catalytic activity. EMBO J. 1997. 16:998–1008.

20. Shigematsu H, Suzuki M, Takahashi T, Miyajima K, Toyooka S, Shivapurkar N, et al. Aberrant methylation of HIN-1 (high in normal-1) is a frequent event in many human malignancies. Int J Cancer. 2005. 113:600–604.

21. Soreide K, Janssen EA, Soiland H, Korner H, Baak JP. Microsatellite instability in colorectal cancer. Br J Surg. 2006. 93:395–406.

22. Asgeirsson KS, Jónasson JG, Tryggvadóttir L, Olafsdóttir K, Sigurgeirsdóttir JR, Ingvarsson S, et al. Altered expression of E-cadherin in breast cancer. Patterns, mechanisms and clinical significance. Eur J Cancer. 2000. 36:1098–1106.
23. Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999. 96:8681–8686.





 PDF
PDF ePub
ePub Citation
Citation Print
Print











 XML Download
XML Download