Abstract
Purpose
The influence of X-inactive specific transcript (XIST) and X-chromosome inactivation associated long non-coding RNAs (lncRNAs) just proximal to XIST (JPX) on hepatocellular carcinoma (HCC) remains controversial in light of previous reports, which the present study aimed to verify.
Materials and Methods
The DIANA lncRNA-microRNA (miRNA) interaction database was used to explore miRNA interactions with JPX or XIST. JPX, XIST, and miR-155-5p expression levels in paired HCC specimens and adjacent normal tissue were analyzed by RT-qPCR. Interaction between XIST and miR-155-5p was verified by dual luciferase reporter assay. Expression levels of miR-155-5p and its known target genes, SOX6 and PTEN, were verified by RT-qPCR and Western blot in HepG2 cells with or without XIST knock-in. The potential suppressive role of XIST and JPX on HCC was verified by cell functional assays and tumor formation assay using a xenograft model.
Results
JPX and XIST expression was significantly decreased in HCC pathologic specimens, compared to adjacent tissue, which correlated with HCC progression and increased miR-155-5p expression. Dual luciferase reporter assay revealed XIST as a direct target of miR-155-5p. XIST knock-in significantly reduced miR-155-5p expression level and increased that of SOX6 and PTEN, while significantly inhibiting HepG2 cell growth in vitro, which was partially reversed by miR-155-5p mimic transfection. JPX knock-in significantly increased XIST expression and inhibited HepG2 cell growth in vitro or tumor formation in vivo in a XIST dependent manner.
The competitive endogenous RNA (ceRNA) network, primarily composed of non-coding RNAs, such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), has now been recognized as an important regulatory network fine-tuning the mRNA and protein expression levels of a vast number of genes in cells. Abnormalities in this network have been found to contribute to the pathogenesis and development of different diseases due to gene dysregulation, the deadliest and perhaps the most heavily studied of which is cancer.12 miRNA regulates gene expression by binding to mRNAs in the complementary base pairing fashion, often at the 3′ un-translated region (3′UTR) of mRNA, leading to cessation of translation and mRNA degradation. lncRNAs primarily reinforce mRNA translation by competitively binding to miRNA targeting mRNA.
Research on the cancer promoting or suppressing roles of miRNAs and lncRNAs in hepatocellular carcinoma (HCC) is still in its beginnings. Several lncRNAs and their interacting miRNAs have been established as promoting or suppressing HCC oncogenesis or progression, such as DANCR, HOTAIR, and HULC.345 However, the functions of other lncRNAs in HCC remain controversial, including the X-chromosome inactivation associated lncRNA X-inactive specific transcript (XIST). Ma, et al.67 reported that down regulation of XIST and another X-associated lncRNA, just proximal to XIST (JPX) were associated with poor prognosis in HCC patients, which is inconsistent with other research implying the HCC suppressing role of these two lncRNA. However, Mo, et al.8 suggested that XIST could promote HCC development by sponging other cancer suppressive miRNAs. The cancer promoting role of XIST has also been reported in non-small cell lung cancer, gastric cancer, and colorectal cancer, while its cancer suppressive function in prostate, breast, and ovarian cancers have also been described.91011121314
The aim of this research was to investigate the function and molecular mechanisms of lncRNA XIST and JPX in HCC. To explore miRNAs that interact with XIST and/or JPX, we surveyed the DIANA lncRNA-miRNA interaction database and found that miR-155-5p, a well-known promoter of HCC development, interacts with XIST.1516 We first examined the expression levels of XIST, JPX, and miR-155-5p in HCC patient specimens and found that expression levels of XIST and JPX inversely correlated with those of miR-155-5p. By dual-luciferase reporter assay and cell functional assays, we confirmed that XIST could suppress HCC by sponging miR-155-5p, abolishing the cancer promoting effect of the latter in HCC, and that the X-chromosome inactivation lncRNA JPX increases XIST expression.
HepG2 human HCC cells were purchased from Thermo Fisher Scientific (Waltham, MA, USA) and cultured in DMEM medium supplemented with 10% FBS and 100 U/mL of penicillin-streptomycin in a humidified incubator with 5% CO2. XIST and JPX knockout, as well as open reading frame knock-in, HepG2 cells were constructed by Genecopoeia (Guangzhou, China). Human XIST shRNA plasmids were purchased from constructs in lentiviral GFP vector from OriGene Technologies (Rockville, MD, USA) and were applied for XIST knockdown following the manufacturer's instructions. miR-155-5p mimic and inhibitor for miR-155-5p overexpression and knockdown, respectively, were purchased from Sigma-Aldrich (Saint Louis, MO, USA) and were applied following the manufacturer's instructions.
This research was approved by the Ethics Review Committee of First Affiliated Hospital of Wenzhou Medical University. Informed consent was obtained from each patient in written form before receiving surgery. HCC pathologic specimens and adjacent tissue were obtained from 20 HCC stage I–II patients and 20 HCC stage III–IV patients. Specimens were preserved both in paraffin blocks and as frozen sections right after surgical resection. Analysis of JPX, XIST, and miR-155-5p expression in tissue specimens was performed as described below. High or low expression was defined as an expression level higher or lower than average, respectively.
Primer pairs for semi-quantifying JPX, XIST, and miR-155-5p expression were purchased as a customized kit from Genecopoeia and applied following the manufacturer's instructions. For JPX and XIST, total RNA was extracted from frozen sections using TRIzol reagent and reversely transcribed into cDNA using a First-Strand cDNA Synthesis Kit (Genecopoeia). For miRNA detection, total RNA was isolated with the same method and reversely transcribed using a miRNA First Strand Synthesis kit (Genecopoeia). Semi-quantification of JPX, XIST, and miR-155-5p was performed by the 2−ΔΔCt method using GAPDH as internal reference for lncRNA and U6 for miRNA. Primer sequences are listed below.
JPX Forward: 5′-TGCAGTCAGAAGGG AGCAAT-3′, Reverse: 5′-CACCGTCATCAGGCTGTCTT-3′. XIST Forward: 5′-CGGGTCTCTTCAAGGACATTTAGCC-3′, Reverse: 5′-GCAC CAATACAGAGGAATG GAGGG-3′. miR-155-5p: 5′-TTAATGC TAATCGTGATAGGGGT-3′.
LncRNA JPX and XIST expression in HCC and adjacent tissue was visualized by in situ hybridization histochemistry (ISHH) using biotin-labeled antisense riboprobes customized by Advanced Cell Diagnostics (Newark, CA, USA) following the manufacturer's instructions. Biotinylated riboprobes were visualized by streptavidin-conjugated horse reddish peroxidase with tetramethylbenzidine as the substrate. Cell nuclei were counterstained with hematoxylin. JPX and XIST expression levels in tissue sections were evaluated as follows: positive staining cells were counted in five randomly-picked microscopic fields on each slide by three different researchers, and relative expression in each slide was determined by normalizing positive staining cell counts to the mean value of that of the adjacent group.
Primary antibodies for Western blot evaluating the protein expression levels of PTEN, SOX6, Bim, Bax, Bcl-2, or beta-actin were previously purchased from Cell Signaling Technology (Danvers, MA, USA). Secondary antibodies previously purchased from Abcam (Cambridge, UK) were used for blotting the primary antibodies. Gray scale analysis was performed using ImageJ (ver 1.52a, National Institutes of Health, Bethesda, MD, USA) software.
Dual luciferase reporter assay was performed using a customized Luc-PairTM Duo-Luciferase Assay Kit purchased from Genecopoeia following the manufacturer's instructions. Briefly, part of XIST cDNA (9350–9400) containing wildtype or mutated putative binding site of miR-155-5p on XIST was cloned into the dual luciferase vector downstream of the firefly luciferase gene. Dual-luciferase vector transfected HepG2 cells were co-transfected with miR-155-5p mimic or inhibitor, and cell lysate was prepared for measuring firefly luciferase activity using Renilla luciferase activity as an internal reference.
Cell viability and proliferation assay was performed using CCK-8 reagent purchased from Dojindo (Shanghai, China) following the manufacturer's instructions. For cell viability assay, cisplatin used for cell treatment was purchased from Tocris Bioscience (Minneapolis, MN, USA) and applied following the manufacturer's instructions. Cell apoptosis was assayed by flow cytometry using an Annexin V Apoptosis Detection Kit purchased from Biolegend (San Diego, CA, USA) following the manufacturer's instructions. Cells with positive Annexin V staining were considered apoptotic cells. Nude mice used for tumor formation assay were purchased from Jackson Laboratory (Bar Harbor, ME, USA) and fed under sterile conditions with unlimited access to standard mouse chow (Harlan) and water. About 5×106 cells were injected into the left back of 4-week-old nude mice, and tumor formation at 1, 2, 3, and 4 week(s) post-injection was evaluated by measuring the tumor mass. Animal use in the present research was approved by the Ethics Review Committee of First Affiliated Hospital of Wenzhou Medical University.
Statistical analysis was performed using Graphpad Prism (ver. 7.0, GraphPad Software, La Jolla, CA, USA). All data represent six replicates and are presented as mean±SD, unless otherwise indicated. Student's t test was used to test significance between one experimental group and one control group, while one-way ANOVA with Dunnett correction was used to test significance between one control group and multiple experimental groups. P<0.05 was considered statistically significant.
To investigate the potential regulatory role of JPX and XIST on miR-155-5p in HCC, we first investigated expression levels of JPX, XIST, and miR-155-5p in 40 pairs of HCC specimens and adjacent non-cancerous specimens previously obtained from 40 patients suffering from HCC, 20 of which were diagnosed with HCC stage I–II and the others with HCC stage III–IV. JPX, XIST, and miR-155-5p expression levels in specimens were analyzed by RT-qPCR; meanwhile, JPX and XIST expression levels in frozen sections were visualized by ISHH. Our results showed that expression levels of XIST and JPX significantly decreased in HCC specimens, compared to those in adjacent tissue, and HCC specimens of TNM stage III–IV showed significant decreases in XIST and JPX expression, compared to that in HCC specimens of TNM stage I–II (Fig. 1A, representatively visualized in Fig. 1C). Expression levels of miR-155-5p were found to be greater in HCC tissue specimens than in adjacent tissue and further increased in HCC tissue of TNM stage III–IV than in HCC tissue specimens of TNM stage I–II (Fig. 1B). The significant inverse correlation in expression levels between miR-155-5p and XIST or JPX in the 40 paired tissue specimens was revealed by correlation analysis (Fig. 1D and E). These data suggested that miR-155-5p is increased in HCC tissue compared to that in adjacent tissue and that miR-155-5p expression is inversely correlated with XIST or JPX in HCC.
By surveying the experimental evidence-based miRNA-lncRNA interaction using the DIANA online database, we found that XIST is a possible target of miR-155-5p. To verify this potential interaction, we performed dual-luciferase reporter assay using HepG2 cells. Plasmid carrying Renilla luciferase cDNA as inner control and firefly luciferase cDNA flanked at 3′ with cDNA inserted with a part of XIST cDNA (9350–9400) containing the wildtype putative miR-155-5p binding site (WT) or mutated counterpart (Mut) were co-transfected with miR-155-5p mimic, miR-155-5p inhibitor, or non-targeting control miRNA in HepG2 cells, which were then measured for firefly/Renilla luciferase activity (Fig. 2A). Renilla luciferase activity remained consistent in four cell groups, although luciferase activity was significantly decreased in HepG2 cells co-transfected with dual-luciferase and miR-155-mimic, compared to the rest of three groups of cells. These effects were abolished when cells were transfected with Mut counterpart (Fig. 2B). These results suggested that XIST might be a direct target of miR-155-5p. To further verify this hypothesis, XIST knock-in (XIST KI) and control (KI NC) HEpG2 cells were constructed and transfected with miR-155-5p mimic or a non-targeting miRNA mimic control. XIST expression was drastically increased by more than three-fold by its knock-in, but significantly decreased in HepG2 cells by miR-155-5p mimic transfection. Meanwhile, miR-155-5p expression was significantly increased by over four-fold by miR-155-5p mimic transfection and significantly decreased by XIST overexpression (Fig. 2C and D). Previous research has suggested that miR-155-5p promotes HCC development by targeting the expression of some tumor suppressor genes, such as SOX6 and PTEN. Therefore, we inspected the protein expression levels of these two genes in HepG2 cells with different XIST or miR-155-5p expression levels. Our results showed that protein expression of SOX6 and PTEN could be significantly increased by XIST overexpression and decreased by miR-155-5p mimic transfection (Fig. 2E–G).
Based on these results, we performed cell functional assays to investigate changes in the expression of XIST or miR-155-5p on cell proliferation, viability, and apoptosis of HepG2 cells. miR-155-mimic transfection significantly increased proliferation of HepG2 cells, consistent with previous reports.1718 XIST overexpression by knocking-in decreased proliferation of HepG2 cells with or without miR-155-5p mimic transfection (Fig. 3A). Cytotoxicity and apoptosis assay revealed that the pro-survival effect of miR-155-5p in HepG2 cells can be antagonized by XIST overexpression: XIST overexpression increased, whereas miR-155-5p mimic transfection decreased, the vulnerability of HepG2 cells to cisplatin treatment, while XIST overexpression did not significantly increase apoptosis in HepG2 cells without cisplatin treatment (Fig. 3B–D). Western blot for detecting the cell apoptosis-related proteins Bim, Bax, and Bcl-2 further confirmed that apoptosis of HepG2 cells with or without miR-155-5p mimic transfection was increased by XIST overexpression when treated with cisplatin (Fig. 3E–G). Together, these results demonstrated that XIST is a direct target of miR-155-5p and that XIST overexpression significantly decreased the cancer promoting effect of miR-155-5p mimic transfection in HepG2 cells in vitro.
Previous research has demonstrated that XIST expression is increased by another X chromosome-associated lncRNA, JPX. Therefore, we investigated whether JPX could regulate miR-155-5p expression and function in HepG2 cells via XIST. JPX knock-in (JPX KI) and JPX KI+XIST knockdown (sh-XIST) in HepG2 cells were constructed. JPX knock-in lifted its expression level by about 3.5-fold, and miR-155-5p mimic transfection or XIST knockdown showed no significant influence on JPX expression. XIST expression was significantly increased by JPX overexpression and decreased by miR-155-5p mimic transfection (Fig. 4A and B). Cell proliferation and apoptosis assay revealed that overexpression of JPX significantly decreased cell proliferation, but increased apoptosis in HepG2 cells, which was abolished by either XIST knockdown or by miR-155-5p mimic transfection (Fig. 4C and D). Western blot for the apoptosis-related proteins yielded similar results (Fig. 4E–G). These data suggested that JPX could decrease cell proliferation, while increasing apoptosis, in HepG2 cells possibly through XIST, which inhibits the function of miR-155-5p.
The molecular mechanism of JPX's tumor suppressing role in HCC or other cancer types remains unclear. Based on our data and previous reports, we speculated that JPX overexpression would suppress HCC development at least in part via increasing increasing XIST expression, and we have verified this hypothesis by performing a tumor formation assay using wild type, JPX knockout, JPX knock-in, and JPX knock-in plus XIST knockdown HepG2 cells. Nude mice injected with JPX-knockout HepG2 cells suffered the most severe tumor growth, suggesting the anti-HCC role of JPX in vivo. JPX overexpression significantly reduced tumor volume growth, which was reduced by XIST knockout, suggesting that the anti-HCC effect of JPX requires the integrity of XIST gene expression (Fig. 5). These data suggested that JPX could repress HCC growth in vivo partially via XIST.
Increasing evidence has shown that miRNAs and lncRNAs are crucial epigenetic regulators in the oncogenesis and HCC progression, most commonly functioning by regulating mRNA translation of key regulatory proteins.19 The contributions of lncRNA JPX and XIST have been well-documented in the X-chromosome inactivation process, balancing the gene expression level between male and female cells.2021 XIST and JPX have also been reported as either cancer promoting or suppressing lncRNA in different cancer types. For example, Xing, et al.22 identified JPX and XIST as suppressive lncRNAs in uveal melanoma, and Yildirim, et al.23 described XIST as a “potent suppressor of hematologic cancer.” Meanwhile, Li, et al.9 reported in a very recent publication that XIST can facilitate non-small cell lung cancer (NSCLC) development by sponging miR-367 and miR-141, thus promoting the epithelial-to mesenchymal transition process in an indirect fashion. Similarly, Jiang, et al.24 suggested that XIST promotes NSCLC progression by sponging miR-137, which targets the cancer promoting gene PXN (paxillin), and Zhang, et al.10 showed that XIST promotes gastric cancer progression by sponging miR-185. However, Zhang, et al.25 demonstrated that XIST could inhibit cell growth and metastasis of osteosarcoma by interacting with miR-21-5p. Current research on the cancer-promoting or -suppressing role of XIST predominantly focuses on its miRNA-interacting potential, and considering that a lncRNA may carry different binding sites for its interaction with different miRNAs, it is not surprising that a given lncRNA may function as either a promotor or suppressor in different cancer types.2627
The impact of changes in XIST and JPX expression levels on HCC currently remains controversial. Downregulation of XIST in HCC pathologic tissue specimens, compared to adjacent non-cancerous tissue, and its association with worsened prognosis in HCC patients were reported by Chang, et al.28 and Ma, et al.7; however, Mo, et al.8 reported a significant increase in XIST expression level in HCC tissue specimens, compared to adjacent tissue, that facilitated the aggressiveness of HCC cells both in vitro and in vivo. Similarly, Ma, et al.67 demonstrated that the downregulation of JPX in HCC patient tissue and plasma, compared to controls, was associated with poor prognosis; however, other researchers reported that JPX levels were greater in HCC patient blood samples than samples from healthy donors.
Aiming to resolve these controversies, we performed current research on the influence of XIST and JPX on HCC cell growth and development. Considering that Sun, et al.29 and Tian, et al.30 have established that JPX could activate XIST transcription by removing the transcription repressor protein CTCF from the XIST promotor and that XIST and JPX often show correlations in their expression level changes,143132 we first presumed that JPX and XIST may have similar influences on HCC progression. We employed the DIANA lncRNA-miRNA indexing database to explore experimentally verified (in both a direct and indirect fashion) miRNAs that target lncRNAs XIST and JPX, and we found that miR-155-5p, whose cancer promoting role in HCC and other cancer types has been heavily investigated, is a potential mutual targeting miRNA of XIST and JPX.17183334 We, therefore, hypothesized that XIST and JPX may suppress HCC cell growth and development by sponging miR-155-5p. However, preliminary results have suggested that miR-155-5p overexpression in various HCC cell lines decreases only the expression of XIST but not JPX. We, hence, primarily focused on XIST-miR-155-5p interactions in HCC in the current research.
By examining HCC pathologic and adjacent tissue, we found that JPX and XIST expression levels were generally decreased in HCC and further decreased with HCC progression, which inversely correlated with miR-155-5p expression level changes. Clinical statistics have revealed a significant correlation between decreases in JPX and XIST or increases in miR-155-5p and HCC progression, implying the suppressing role of JPX and XIST, as well as the promoting role of miR-155-5p, in HCC. These results are consistent with previous reports.7283536 By dual luciferase reporter assay and examining XIST expression status in XIST overexpressing HepG2 cells transfected with miR-155-5p mimic, we identified XIST as a direct target of miR-155-5p, and overexpressing XIST was able to partially rescue the protein expression levels of some HCC suppressing genes previously identified as direct targets of miR-155-5p in miR-155-5p mimic-transfected HepG2 cells, including PTEN and SOX6.161837 The HCC suppressing role of XIST antagonizing that of miR-155-5p was further revealed by cell functional assays: XIST overexpression not only decreased cell proliferation while increasing apoptosis, but also increased the susceptibility of HepG2 cells to cisplatin treatment, which was undermined by miR-155-5p mimic transfection. Moreover, the results of our cell functional assays using JPX overexpressing HepG2 cells revealed that JPX may suppress HCC development via XIST. JPX overexpression significantly increased XIST expression level, which was abolished by miR-155-5p transfection. JPX overexpression also significantly inhibited cell proliferation and in vivo tumor formation while increasing apoptosis in HepG2 cells, which was reduced by either XIST knockdown or miR-155-5p transfection.
Collectively, our results suggested that JPX may suppress HCC cell growth and development by increasing the expression levels of XIST, which functions as a suppressor of HCC by sponging the cancer promoting miR-155-5p. These results further emphasized the prognostic and therapeutic value of XIST and JPX, providing new insights on the cancer suppressing role of these two lncRNAs. Interestingly, another X-chromosome inactivation associated lncRNA, FTX, has also been reported as a HCC suppressor. Expression levels of FTX and XIST have both been found to be higher in females than that in males, which may be related to the gender difference of HCC susceptibility.3839 Currently, the impact of X-inactivation disorder on HCC has not been explored in detail, although abnormality in X chromosome inactivation has been linked with BRCA1/2 inactivation in breast and ovarian cancer.4041 Further research on the oncogenic mechanism of disorders in X chromosome inactivation is warranted.
Figures and Tables
Fig. 1
XIST and JPX expression levels inversely correlated with HCC development and miR-155-5p expression level. (A and B) Expression levels of XIST and JPX significantly decreased in 20 HCC tissue specimens compared to adjacent counterparts and further decreased in advanced staged HCC tissue specimens, compared to early staged ones, while increases in miR-155-5p expression are significantly correlated with HCC oncogenesis and progression. Data were normalized to the mean of adjacent tissue and are presented as fold changes. (C) Representative images of JPX and XIST hybridization histochemistry in situ (magnification, ×200). Long non-coding RNAs in the frozen section were hybridized with biotin-labeled riboprobes in a complementary base pairing manner (pale brown). Cell nuclei were counterstained with hematoxylin (cyan). (D and E) Correlation analysis showing that expression levels of JPX and XIST are inversely correlated with those of miR-155-5p in all 20 pairs of HCC and adjacent tissue. Relative expression in (A) and (B) was used for correlation analysis in (D) and (E). *p<0.05, †p<0.01, ‡p<0.001. XIST, X-inactive specific transcript; JPX, just proximal to XIST; HCC, hepatocellular carcinoma.
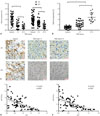
Fig. 2
XIST is a direct target of miR-155-5p. (A) Demonstration of XIST cDNA inserted into the 3′UTR of firefly luciferase gene with putative binding sites of miR-155-5p. (B) Dual luciferase reporter assay results suggesting that miR-155-5p directly targets XIST. (C and D) XIST expression was significantly increased by knocking in and decreased by miR-155-5p mimic transfection in wild type (knock-in control) and XIST knock-in HepG2 cells. (E and F) Western blot results showing that protein expression levels of previously identified miR-155-5p targeting genes PTEN and SOX6 were significantly decreased by miR-155-5p mimic transfection and partially rescued by XIST knock-in in HepG2 cells. Semi-quantification of protein analysis was performed by comparing the gray scale of PTEN or SOX6 protein bands to that of β-actin. (G) Representative results of Western blots evaluating PTEN and SOX6 expression in HepG2 cells with different miR-155-5p and XIST expression statuses. Data are presented as fold changes compared to the mean of a control group (presented as the first bar on the left of each graph). *p<0.05, †p< 0.01, ‡p<0.001. XIST, X-inactive specific transcript; JPX, just proximal to XIST; KI NC, knock-in control; XIST KI, XIST knock-in.

Fig. 3
XIST suppresses cell growth while inducing apoptosis in HepG2 cells by antagonizing the cancer promoting effect of miR-155-5p. Cells were treated with cisplatin at indicated concentrations for 24 h before analysis. (A) Cell proliferation assay showing that increase in cell proliferation rate induced by miR-155-5p mimic transfection can be repressed by XIST knock-in. (B–D) XIST knock-in significantly reduced cell viability while increasing apoptosis and cisplatin susceptibility of HepG2 cells with or without miR-155-5p mimic transfection. (C) is a representative result of flow cytometry counting for the percentage of Annexin V positive cells in each group. (E–G) miR-155-5p mimic transfection decreased, while XIST knock-in increased, apoptosis-related protein parameters in HepG2 cells with or without cisplatin treatment. (G) is a representative result of Bim, Bax, and Bcl-2 protein expression levels in different groups. Semi-quantitative analysis of Western blot results was performed as described in Fig. 2. Western blot data were presented as fold-changes compared to that of a control group. *p<0.05, †p< 0.01, ‡p<0.001, §p<0.0001. XIST, X-inactive specific transcript; JPX, just proximal to XIST; KI NC, knock-in control; XIST KI, XIST knock-in.
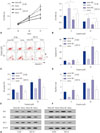
Fig. 4
JPX suppresses HepG2 cell growth while inducing apoptosis in HepG2 cells via XIST. (A and B) JPX expression levels were significantly increased by knocking in. XIST expression levels were significantly increased by JPX knock-in. Meanwhile, miR-155-5p mimic transfection did not alter JPX expression levels. (C and D) The anti-proliferation and apoptosis-inducing mechanism of JPX in HepG2 cells requires the integrity of XIST. (E–G) Results of Western blot analysis on apoptosis-related protein parameters correlated with cell apoptosis in graph (D). Semi-quantitative analysis of Western blot results was performed as described in Fig. 2. Western blot data are presented as fold-changes compared to that of controls. *p<0.05, †p< 0.01, ‡p<0.001. XIST, X-inactive specific transcript; JPX, just proximal to XIST; JPX KI, JPX knock-in; KI NC, knock-in control; sh-XIST, XIST knockdown.

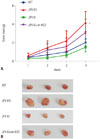
Fig. 5
Tumor formation in nude mice reveals the HCC suppressive role of XIST and JPX. Twelve nude mice were randomly assigned to each group for cell injection on the left side of the back. (A) Tumor mass was weighed at 1, 2, 3, and 4 week(s) post-injection. (B) is a representative result of tumor formation at week 4. Statistical analysis was performed on tumor weight at week 4. *p<0.01. HCC, hepatocellular carcinoma; XIST, X-inactive specific transcript; JPX, just proximal to XIST.
ACKNOWLEDGEMENTS
This study was funded by a grant from the Natural Science Foundation of China (No. 81572780).
References
1. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014; 505:344–352.

2. Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013; 3:1113–1121.

3. Yuan SX, Wang J, Yang F, Tao QF, Zhang J, Wang LL, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016; 63:499–511.

4. Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang H, et al. Hotair mediates hepatocarcinogenesis through suppressing miRNA-218 expression and activating P14 and P16 signaling. J Hepatol. 2015; 63:886–895.

5. Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, et al. Long non-coding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015; 75:846–857.

6. Ma X, Yuan T, Yang C, Wang Z, Zang Y, Wu L, et al. X-inactive-specific transcript of peripheral blood cells is regulated by exosomal Jpx and acts as a biomarker for female patients with hepatocellular carcinoma. Ther Adv Med Oncol. 2017; 9:665–677.

7. Ma W, Wang H, Jing W, Zhou F, Chang L, Hong Z, et al. Downregulation of long non-coding RNAs JPX and XIST is associated with the prognosis of hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2017; 41:163–170.

8. Mo Y, Lu Y, Wang P, Huang S, He L, Li D, et al. Long non-coding RNA XIST promotes cell growth by regulating miR-139-5p/PDK1/AKT axis in hepatocellular carcinoma. Tumour Biol. 2017; 39:1010428317690999.

9. Li C, Wan L, Liu Z, Xu G, Wang S, Su Z, et al. Long non-coding RNA XIST promotes TGF-β-induced epithelial-mesenchymal transition by regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer. Cancer Lett. 2018; 418:185–195.

10. Zhang Q, Chen B, Liu P, Yang J. XIST promotes gastric cancer (GC) progression through TGF-β1 via targeting miR-185. J Cell Biochem. 2018; 119:2787–2796.

11. Du Y, Weng XD, Wang L, Liu XH, Zhu HC, Guo J, et al. LncRNA XIST acts as a tumor suppressor in prostate cancer through sponging miR-23a to modulate RKIP expression. Oncotarget. 2017; 8:94358–94370.

12. Xiao C, Sharp JA, Kawahara M, Davalos AR, Difilippantonio MJ, Hu Y, et al. The XIST noncoding RNA functions independently of BRCA1 in X inactivation. Cell. 2007; 128:977–989.

13. Liu X, Cui L, Hua D. Long non-coding RNA XIST regulates miR-137-EZH2 axis to promote tumor metastasis in colorectal cancer. Oncol Res. 2018; 03. 01. [Epub]. DOI: 10.3727/096504018X15195193936573.

14. Huang YS, Chang CC, Lee SS, Jou YS, Shih HM. Xist reduction in breast cancer upregulates AKT phosphorylation via HDAC3-mediated repression of PHLPP1 expression. Oncotarget. 2016; 7:43256–43266.

15. Yan XL, Jia YL, Chen L, Zeng Q, Zhou JN, Fu CJ, et al. Hepatocellular carcinoma-associated mesenchymal stem cells promote hepatocarcinoma progression: role of the S100A4-miR155-SOCS1-MMP9 axis. Hepatology. 2013; 57:2274–2286.

16. Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, Patel T, et al. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009; 50:1152–1161.

17. Chen G, Wang D, Zhao X, Cao J, Zhao Y, Wang F, et al. miR-155-5p modulates malignant behaviors of hepatocellular carcinoma by directly targeting CTHRC1 and indirectly regulating GSK-3β-involved Wnt/β-catenin signaling. Cancer Cell Int. 2017; 17:118.

18. Fu X, Wen H, Jing L, Yang Y, Wang W, Liang X, et al. MicroRNA-155-5p promotes hepatocellular carcinoma progression by suppressing PTEN through the PI3K/Akt pathway. Cancer Sci. 2017; 108:620–631.

19. Wong CM, Tsang FH, Ng IO. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol. 2018; 15:137–151.

20. Pintacuda G, Young AN, Cerase A. Function by structure: spotlights on Xist long non-coding RNA. Front Mol Biosci. 2017; 4:90.

21. Robert Finestra T, Gribnau J. X chromosome inactivation: silencing, topology and reactivation. Curr Opin Cell Biol. 2017; 46:54–61.

22. Xing Y, Wen X, Ding X, Fan J, Chai P, Jia R, et al. CANT1 lncRNA triggers efficient therapeutic efficacy by correcting aberrant lncing cascade in malignant uveal melanoma. Mol Ther. 2017; 25:1209–1221.

23. Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013; 152:727–742.

24. Jiang H, Zhang H, Hu X, Li W. Knockdown of long non-coding RNA XIST inhibits cell viability and invasion by regulating miR-137/PXN axis in non-small cell lung cancer. Int J Biol Macromol. 2018; 111:623–631.

25. Zhang R, Xia T. Long non-coding RNA XIST regulates PDCD4 expression by interacting with miR-21-5p and inhibits osteosarcoma cell growth and metastasis. Int J Oncol. 2017; 51:1460–1470.

26. Lin C, Yang L. Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol. 2018; 28:287–301.

27. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017; 77:3965–3981.

28. Chang S, Chen B, Wang X, Wu K, Sun Y. Long non-coding RNA XIST regulates PTEN expression by sponging miR-181a and promotes hepatocellular carcinoma progression. BMC Cancer. 2017; 17:248.

29. Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. Jpx RNA activates Xist by evicting CTCF. Cell. 2013; 153:1537–1551.

30. Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010; 143:390–403.

31. Romito A, Rougeulle C. Origin and evolution of the long non-coding genes in the X-inactivation center. Biochimie. 2011; 93:1935–1942.

32. Chow JC, Hall LL, Clemson CM, Lawrence JB, Brown CJ. Characterization of expression at the human XIST locus in somatic, embryonal carcinoma, and transgenic cell lines. Genomics. 2003; 82:309–322.

33. Paraskevopoulou MD, Vlachos IS, Karagkouni D, Georgakilas G, Kanellos I, Vergoulis T, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016; 44:D231–D238.

34. Wang M, Yang F, Qiu R, Zhu M, Zhang H, Xu W, et al. The role of mmu-miR-155-5p-NF-κB signaling in the education of bone marrow-derived mesenchymal stem cells by gastric cancer cells. Cancer Med. 2018; 7:856–868.

35. Zhang Y, Wei C, Guo CC, Bi RX, Xie J, Guan DH, et al. Prognostic value of microRNAs in hepatocellular carcinoma: a meta-analysis. Oncotarget. 2017; 8:107237–107257.

36. Zhang L, Wang W, Li X, He S, Yao J, Wang X, et al. MicroRNA-155 promotes tumor growth of human hepatocellular carcinoma by targeting ARID2. Int J Oncol. 2016; 48:2425–2434.

37. Xie Q, Chen X, Lu F, Zhang T, Hao M, Wang Y, et al. Aberrant expression of microRNA 155 may accelerate cell proliferation by targeting sex-determining region Y box 6 in hepatocellular carcinoma. Cancer. 2012; 118:2431–2442.

38. Liu F, Yuan JH, Huang JF, Yang F, Wang TT, Ma JZ, et al. Long non-coding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene. 2016; 35:5422–5434.

39. Zhuang LK, Yang YT, Ma X, Han B, Wang ZS, Zhao QY, et al. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 2016; 7:e2203.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download