Abstract
Aspirin-induced asthma (AIA) and aspirin-induced urticaria/angioedema (AIU) are two major aspirin-related allergies. We summarize recent findings related to their molecular genetic mechanisms in order to identify genetic susceptibility markers for differentiating AIU and AIA. The overproduction of cysteinyl leukotriene has been suggested as a mechanism in both AIU and AIA. Increased expression of CYSLTR1 with CYLSTR1 and CYSLTR2 polymorphisms are new findings in AIA, while the ALOX5 promoter polymorphism has been noted in AIU. An HLA study suggested that DPB1*0301 is a strong genetic marker for AIA, and that HLA DRB1*1302 and DQB1*0609 are markers for AIU susceptibility. Several single nucleotide polymorphisms (SNPs) in the promoters of EP2, TBX21, COX-2, FcεRIβ, and TBXA2R were associated with AIA, while an FcεRIα promoter polymorphism was associated with AIU. The functional studies of the key genes involved in AIA and AIU are summarized. The identification and functional study of genetic markers for AIA and AIU susceptibility would further elucidate the pathogenic mechanisms and facilitate the development of early diagnostic markers to establish therapeutic targets.
Aspirin (acetyl salicylic acid) ingestion induces several allergic diseases, including aspirin-intolerant asthma (AIA), aspirin-induced acute or chronic urticaria/angioedema (AIAU or AICU), anaphylaxis, and rarely, hypersensitivity pneumonitis.1,2 Of these conditions, AIA and AIU are the most prevalent in Korea. Although the pathogenic mechanism of AIA is not completely understood, the chronic overproduction of cysteinyl leukotrienes (Cys-LTs) derived from cyclooxygenase (COX) inhibition is a consistent finding.3-6 In contrast to AIA, information on the pathogenic mechanism of AIU is limited. Recent reports, however, have suggested that Cys-LT overproduction is one pathogenic mechanism, which is also associated with AIU.7-9 To explore the molecular genetic mechanisms of AIA and AIU, we performed several case-control studies to identify gene markers for differentiating AIA and AIU, focusing on HLA alleles and leukotriene-related genes.
AIA is characterized by a typical sequence of symptoms, including intense eosinophilic inflammation of the upper and lower airways. It is more commonly found in non-atopic, middle-aged females and is strongly associated with chronic rhinosinusitis and nasal polyposis.10 Typically, the asthmatic symptoms can be classified as moderate to severe, and long-term maintenance medications, including leukotriene-receptor antagonists, are required to control the asthma symptoms.
Aspirin ingestion can induce acute and chronic urticaria. Chronic urticaria is defined as suffering from daily urticaria symptoms for more than 6 weeks.7 AIU can be confirmed by the oral aspirin challenge test.11,12 Based on these test results, chronic urticaria patients are classified into two groups: those with chronic urticaria associated with aspirin sensitivity (AICU) and those with negative responses (chronic idiopathic urticaria, CIU). In our previous study,13 AICU patients (compared to CIU patients) were found to be younger and showed higher rates of atopy and serum total IgE. No significant differences were noted in the prevalence of thyroid autoantibodies, antinuclear antibody, or other clinical parameters.
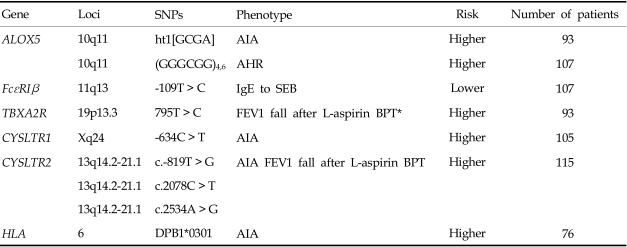
The association between HLA-DPB1*0301 and AIA was first reported in a Polish population,14 and the results were replicated in a Korean population.15 The frequency of DPB1*0301 was significantly higher in AIA patients compared to normal and asthmatic controls, suggesting that immune system recognition of an unknown antigen is part of the pathogenesis of AIA.14 In the Korean population,15 the patients with DPB1*0301 tended to have lower FEV1 levels, be female, and have a higher prevalence of rhinosinusitis and nasal polyps than those without it. These are the typical clinical symptoms of AIA. Furthermore, the presence of HLA-DPB1*0301 was significantly associated with a higher leukotriene receptor antagonist dose requirement in the long-term management of AIA patients.16 We postulate that HLA-DPB1*0301 is a strong genetic marker for identifying the AIA phenotype.
It has been suggested that the activity of the leukotriene C4 synthase gene (LTC4S), a key enzyme in Cys-LT synthesis, is genetically regulated in the pathogenesis of AIA. Two promoter polymorphisms (-1072G > A and -444A > C) have been identified, and LTC4S-444A > C was reported to be positively associated with the AIA phenotype in a Polish population.17,18 The patients with the C allele had a greater risk of developing AIA. The adenosine to cytosine conversion of this promoter polymorphism created an additional responsive element for histone H4 transcription factor-2, increasing the transcription rate of the gene both in vitro and in vivo. Patients carrying the LTC4S-444A homozygote had a greater increase in urinary LTE4 after an aspirin challenge.19 The association of this promoter polymorphism with the phenotype of AIA patients, however, has not been replicated in other populations, including Japanese, American, and Korean populations.19-21
Some reports have suggested the possible involvement of the 5-lipoxygenase gene (ALOX5) in AIA. The Drazen group first identified the promoter polymorphism of the ALOX5 gene, which consisted of a variable number of tandem repeats of GC-rich motifs associated with the binding of Sp1 transcription factors.22 Subjects with the wild-type genotype (five repeats) had a significantly higher capacity to produce Cys-LTs compared to those with a mutant genotype (3, 4, or 6 repeats). Furthermore, a significant association between the ALOX5 promoter polymorphism and the severity of airway hyperresponsiveness was detected in the Korean population.23 AIA patients carrying a mutant allele (n ≠ 5 repeats) showed increased airway hyperresponsiveness compared to AIA patients with the wild-type genotype. When we screened ten SNPs for key enzymes involved in arachidonate metabolism in a Korean population, 5-lipoxygenase (ALOX5, -1708G > A, 21C > T, 270G > A, 1728G > A), ALOX5-activating protein (ALOX5AP, 218A > G), cyclooxygenase 2 (COX-2, -162C > G, 10T > G, 228G > A), leukotriene C4 synthase (LTC4S, -444A > C), and cysteinyl leukotriene receptor 1 (CYSLTR1, 927T > C), the lack of an association was noted between the ALOX5AP, COX-2, and CYSLTR1 gene polymorphisms and the AIA phenotype.21 The possible involvement of ALOX5-ht1 [G-C-G-A] of the ALOX5 gene (-1708G > A, 21C > T, 270G > A, 1728G > A) in AIA development, however, was suggested because the frequency of the [G-C-G-A] haplotype of ALOX5 in the AIA group was significantly higher than its frequency in the ATA group.21
Cys-LTs exert their biological actions by binding to two types of receptors: CYSLTR1 and CYSLTR2. We have found genetic associations between these two genes and AIA.24,25 Three SNPs (-634C > T, -475A > C, -336A > G) in the 5'-upstream region of CYSLTR124 were significantly associated with the AIA phenotype, especially in males. Male AIA patients had higher frequencies of the minor alleles, while no differences were noted in females. Furthermore, these promoter polymorphisms were associated with increased promoter activity. A functional study using two cell lines, human T lymphocytes (Jurkat) and lung epithelial cells (A549), showed enhanced promoter activity with the ht2 [T-C-G] construct compared to the ht1 [C-A-A] construct, suggesting that this polymorphism modulates CYSLTR1 expression to increase AIA susceptibility. Four SNPs of the CYSLTR2 polymorphism, c.-819T > G, c.2078C > T, c.2534A > G, and c.2545 +297A > G, were found in a Korean population25 and the frequencies of the rare alleles c.-819T > G, c.2078C > T, and c.2534A > G were higher in those with AIA than in those with ATA. Patients with the rare alleles c.-819T > G, c.2078C > T, or c.2534A > G exhibited a greater percentage drop in FEV1 after aspirin provocation than those with the common allele. These results suggest that CYSLTR1 and ALOX5, but not LTC4S, can be genetic markers for AIA susceptibility in the Korean population.
In a case-controlled study of 63 candidate genes in a Japanese population,26 a functional SNP of the PGE2 receptor subtype 2 gene (EP2) was found to be associated with the risk of AIA. This SNP may act by reducing the PGE2 braking mechanism of inflammation. Although a novel promoter polymorphism (-765G > C) of COX-2 was not associated with AIA, the CC homozygote of this promoter polymorphism was associated with increased PGE2 production by creating an E2F transcription factor binding motif.27 Further studies are needed to clarify the role of COX-2 and PG imbalance in the pathogenic mechanisms of AIA in a Korean population.
TBXA2R is a receptor for a potent bronchoconstrictor, thromboxane A2 (TBXA2), and a TBXA2R + 795T > C polymorphism could augment the bronchoconstrictive response to inhaled aspirin, possibly contributing to AIA development in the Korean population.28 The effect of increased TBXA2 production in the pathogenesis of AIA could be explained by the fact that oral aspirin administration uncouples the TBXA2-dependent negative feedback mechanisms, which may, in turn, increase the production of Cys-LTs.29 The TBXA2-dependent regulation of LTC4S activity may be an important pathophysiological mechanism in AIA.
We investigated two polymorphisms of FcεR1β (-109T > C and E237G) in a Korean population and found no significant association between the genotype distributions of these polymorphisms and the phenotype of AIA.30 However, a significant association between the -109T > C polymorphism and IgE specific to staphylococcal enterotoxin B (SEB) was found. The number of subjects with both homozygous alleles of FcεR1β-109T and IgE specific to SEB was significantly greater in the AIA group. Furthermore, luciferase reporter assays showed that the FcεR1β-109T allele had higher promoter activity in both the RBL-2H3 and A549 cell lines, suggesting that the FcεR1β-109T > C polymorphism increases the expression of FcεR1β in mast cells. This may lead to the enhanced release of proinflammatory mediators in the asthmatic airway, and thereby contributing to increased AIA susceptibility.
TBX21 encodes T-bet, a T-box transcription factor expressed in T cells, which influences naive T lymphocyte development and has been implicated in asthma pathogenesis.31 The -1993T T > C SNP in the TBX21 promoter was found to be associated with an increased risk for AIA in a Japanese population by increasing transcriptional activity.32 This genetic variation can cause inappropriate Th1 responses in the airway, leading to severe airway inflammation in combination with antigen-specific Th2 responses. That report concluded that the Th1 response played a role in the pathogenesis of AIA equal to that of the Th2 response.
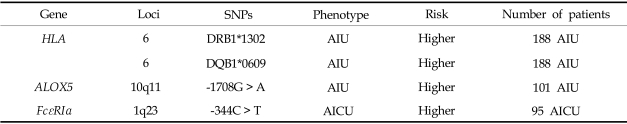
We demonstrated an association between the HLA genotype and AIU using a high-resolution analysis technique and confirmed a strong positive association between the HLA-DRB1*1302 and HLA-DQB1*0609 alleles in AIU patients.33 The allele frequencies of HLA-DRB1*1302 and HLA-DQB1*0609 in AIU patients were significantly higher than in AIA patients and normal controls (NC). In this study, we suggested that HLA-DRB1*1302-DQB1*0609-DPB1*0201 is a strong genetic determinant of the AIU phenotype. This is the first study to demonstrate a positive association between an HLA allele and the AIU phenotype. It suggests that among patients suffering from aspirin- or NSAID-related allergies, subjects carrying the HLA-DPB1*0301 allele may represent the AIU phenotype, while those carrying HLA-DRB1*1302 or HLA-DQB1*0609 may represent the AIU phenotype.
Although there is no clear evidence of the involvement of Cys-LTs in AIU, it has been proposed that Cys-LTs participate in the pathogenesis of urticaria and angioedema, by a mechanism similar to that in AIA. Mastalerz et al. demonstrated the overproduction of Cys-LT in association with the LTC4S polymorphism in chronic urticaria patients with aspirin sensitivity.34 The frequency of subjects carrying the C allele of LTC4S-444A > C was significantly higher in chronic urticaria patients who tested positive in the aspirin challenge test than those who tested negative. No association, however, was found between the LTC4S-444A > C polymorphism and the phenotype of NSAID-induced isolated periorbital angioedema in Spanish35 or Korean populations.36 We also investigated the genetic polymorphisms of candidate genes encoding enzymes which are involved in leukotriene synthesis in a Korean population.36 Of the eight SNPs in the four leukotriene-related genes, ALOX5, ALOX5AP (5-lipoxygenase activating protein), and PTGS2 (cyclooxygenase 2), as well as the polymorphism of ALOX5 at position-1708 G > A, showed a significant difference in genotypic frequency between AIU and AIA. The frequency of the ALOX5-1708A allele in the AIU group was significantly lower than that in the AIA group. No significant differences were noted in the other SNPs, suggesting that ALOX5 makes different contributions in AIU and AIA, and in AIU and NC.
Considering the pathogenic mechanisms of AIU, we postulate that the polymorphisms of candidate genes involved in histamine release and metabolism and their receptors (H1 and H2 receptors) are associated with the AIU phenotype. First, we analyzed 11 known SNPs of the high-affinity IgE receptor I (FcεRIβ), histamine N-methyl transferase (HNMT), histamine receptor type 1 (HRH1), histamine receptor type 2 (HRH2), and their haplotypes in AIU patients, and compared them to other drug allergy patients and normal controls. No significant differences were observed in allele or genotype frequencies, and the haplotype frequencies of all SNPs of FcεRIβ, HNMT, HRH1, and HRH2 were similar in the three groups.37 This finding suggests that polymorphisms of FcεRIβ and the three histamine-related genes do not contribute to the development of the AIU phenotype in the Korean population. We also investigated two genetic polymorphisms (-344C > T, -95 T > C) of the FcεRIα gene in patients with aspirin-induced chronic urticaria (AICU), and compared them to patients with aspirin-tolerant chronic urticaria (ATCU) and NC.38 Of the two genetic polymorphisms of FcεRIα, AICU patients had a significantly higher frequency of the genotype containing the -344T allele than the ATCU and NC groups. No significant differences were noted in the genotype or allele frequencies of the -95T > C polymorphism among the three groups. In addition, the genetic variants of FcεRIα -344C > T appeared to increase promoter activity in both the A549 and RBL-2H3 cell lines. These findings suggest that the FcεRIα -335C > T polymorphism contributes to the development of AICU by increasing the promoter activity. Further investigation is needed to elucidate whether this genetic variant can increase histamine releasability or augment the signaling from mast cells, which leads to the release of proinflammatory mediators. Table 2 summarizes the gene lists representing the phenotype of AIU.
The diagnosis of aspirin sensitivity can be made with certainty using challenge tests involving increasing doses of aspirin. There are four types of challenge tests based on the route of aspirin administration: oral, bronchial, nasal, and rarely, intravenous tests.11,12 Both the oral and bronchial tests have similar specificities, although the oral test has somewhat greater sensitivity. Because the oral aspirin challenges can induce severe reactions and take several days to complete, the lysine-aspirin bronchial challenge test is widely used in European and Asian countries.12 As an in vitro test, the measurement of 15-HETE induced from peripheral blood by aspirin has been suggested as a specific test for aspirin-sensitive patient identification (ASPI test).39
When aspirin sensitivity is confirmed in both AIA and AIU patients, the complete avoidance of aspirin/NSAIDs is crucial to prevent life-threatening, adverse reactions. Moreover, the avoidance of salicylate-containing foods is recommended. The basic principle of pharmacotherapy for AIA is step-wise treatment based on anti-inflammatory medications. A leukotriene receptor antagonist (LTRA) is the first line agent for AIA,40-42 and LTRA has a beneficial effect in rhinitis/nasal polyposis. Aspirin desensitization may reduce the inflammatory mucosal symptoms of the upper and lower airways. Antihistamines are the first-line pharmacotheraputic treatment for AIU patients. LTRAs also have a beneficial effect.43
Further information on the genetic polymorphisms of these candidate genes with supporting functional studies will make it possible to elucidate the molecular mechanisms of two major aspirin-related conditions, AIA and AIU, and should lead to the identification of genetic susceptibility markers for differentiating AIA and AIU. This will lead to the development of new diagnostic markers and provide additional therapeutic targets based on genetic information.
References
1. Simon RA. Adverse respiratory reactions to aspirin and nonsteroidal anti-inflammatory drugs. Curr Allergy Asthma Rep. 2004; 4:17–24. PMID: 14680616.

2. Szczeklik A, Stevenson DD. Aspirin-induced asthma: advances in pathogenesis, diagnosis, and management. J Allergy Clin Immunol. 2003; 111:913–921. PMID: 12743549.

3. Pierzchalska M, Mastalerz L, Sanak M, Zazula M, Szczeklik A. A moderate and unspecific release of cysteinyl leukotrienes by aspirin from peripheral blood leucocytes precludes its value for aspirin sensitivity testing in asthma. Clin Exp Allergy. 2000; 30:1785–1791. PMID: 11122218.

4. Antczak A, Montuschi P, Kharitonov S, Gorski P, Barnes PJ. Increased exhaled cysteinyl-leukotrienes and 8-isoprostane in aspirin-induced asthma. Am J Respir Crit Care Med. 2002; 166:301–306. PMID: 12153961.

5. Christie PE, Tagari P, Ford-Hutchinson AW, Charlesson S, Chee P, Arm JP, et al. Urinary leukotriene E4 concentrations increase after aspirin challenge in aspirin-sensitive asthmatic subjects. Am Rev Respir Dis. 1991; 143:1025–1029. PMID: 1850964.
6. Oosaki R, Mizushima Y, Mita H, Shida T, Akiyama K, Kobayashi M. Urinary leukotriene E4 and 11-dehydrothromboxane B2 in patients with aspirin-sensitive asthma. Allergy. 1997; 52:470–473. PMID: 9188934.
7. Grattan CE. Aspirin sensitivity and urticaria. Clin Exp Dermatol. 2003; 28:123–127. PMID: 12653694.

8. Mastalerz L, Setkowicz M, Sanak M, Szczeklik A. Hypersensitivity to aspirin: Common eicosanoid alterations in urticaria and asthma. J Allergy Clin Immunol. 2004; 113:771–775. PMID: 15100686.

9. Sanchez-Borges M, Capriles-Hulett A, Caballero-Fonseca F. Cutaneous reactions to aspirin and nonsteroidal anti-inflammatory drugs. Clin Rev Allergy Immunol. 2003; 24:125–136. PMID: 12668893.
10. Szczeklik A, Nizankowska E, Duplaga M. Natural history of aspirin-induced asthma. AIANE Investigators. European Network on Aspirin-Induced Asthma. Eur Respir J. 2000; 16:432–436. PMID: 11028656.
11. Park HS. Early and late onset asthmatic responses following lysine-aspirin inhalation in aspirin-sensitive asthmatic patients. Clin Exp Allergy. 1995; 25:38–40. PMID: 7728622.

12. Nizankowska E, Bestynska-Krypel A, Cmiel A, Szczeklik A. Oral and bronchial provocation tests with aspirin for diagnosis of aspirin-induced asthma. Eur Respir J. 2000; 15:863–869. PMID: 10853850.

13. Ye YM, Kim JE, Kim SH, Suh CH, Nahm DH, Park HS. Clinical characteristics of chronic urticaria with aspirin sensitivity. Korean J Asthma Allergy Clin Immunol. 2005; 25:194–199.
14. Dekker JW, Nizankowska E, Schmitz-Schumann M, Pile K, Bochenek G, Dyczek A, et al. Aspirin-induced asthma and HLA-DRB1 and HLA-DPB1 genotypes. Clin Exp Allergy. 1997; 27:574–577. PMID: 9179433.

15. Choi JH, Lee KW, Oh HB, Lee KJ, Suh YJ, Park CS, et al. HLA association in aspirin-intolerant asthma: DPB1*0301 as a strong marker in a Korean population. J Allergy Clin Immunol. 2004; 113:562–564. PMID: 15007363.
16. Park HS, Kim SH, Sampson AP, Lee KW, Park CS. The HLA-DPB1*0301 marker might predict the requirement for leukotriene receptor antagonist in patients with aspirin-intolerant asthma. J Allergy Clin Immunol. 2004; 114:688–689. PMID: 15446291.

17. Sanak M, Simon HU, Szceklik A. Leukotriene C4 synthase promoter polymorphism and risk of aspirin-induced asthma. Lancet. 1997; 350:1599–1600. PMID: 9393345.

18. Sanak M, Pierzchalska M, Bazan-Socha S, Szczeklik A. Enhanced expression of the leukotriene C4 synthase due to overactive transcription of an allelic variant associated with aspirin-intolerant asthma. Am J Respir Cell Mol Biol. 2000; 23:290–296. PMID: 10970818.
19. Kawagishi Y, Mita H, Taniguchi M, Maruyama M, Oosaki R, Higashi N, et al. Leukotriene C4 synthase promoter polymorphism in Japanese patients with aspirin-induced asthma. J Allergy Clin Immunol. 2002; 109:936–942. PMID: 12063521.

20. Van Sambeek R, Stevenson DD, Baldasaro M, Lam BK, Zhao J, Yoshida S, et al. 5' Flanking region polymorphism of the gene encoding leukotriene C4 synthase does not correlate with the aspirin-intolerant asthma phenotype in the United States. J Allergy Clin Immunol. 2000; 106:72–76. PMID: 10887308.

21. Choi JH, Park HS, Oh HB, Lee JH, Suh YJ, Park CS, et al. Leukotriene-related gene polymorphisms in ASA-intolerant asthma: an association with a haplotype of 5-lipoxygenase. Hum Genet. 2004; 114:337–344. PMID: 14749922.

22. In KH, Asano K, Beier D, Grobholz J, Finn PW, Silverman EK, et al. Naturally occurring mutations in the human 5-lipoxygenase gene promoter that modify transcription factor binding and reporter gene transcription. J Clin Invest. 1997; 99:1130–1137. PMID: 9062372.

23. Kim SH, Bae JS, Suh CH, Nahm DH, Holloway JW, Park HS. Polymorphism of tandem repeat in promoter of 5-lipoxygenase in ASA-intolerant asthma: a positive association with airway hyperresponsiveness. Allergy. 2005; 60:760–765. PMID: 15876305.

24. Kim SH, Park HS, Choi JH, Oh JM, Suh CH, Nahm DH. A strong association of three genetic variants of CYSLTR1 with ASA-intolerant asthma phenotype. J Allergy Clin Immunol. 2005; 115:S134.

25. Park JS, Chang HS, Park CS, Lee JH, Lee YM, Choi JH, et al. Association analysis of cysteinyl-leukotriene receptor 2 (CYSLTR2) polymorphisms with aspirin intolerance in asthmatics. Pharmacogenet Genomics. 2005; 15:483–492. PMID: 15970796.

26. Jinnai N, Sakagami T, Sekigawa T, Kakihara M, Nakajima T, Yoshida K, et al. Polymorphisms in the prostaglandin E2 receptor subtype 2 gene confer susceptibility to aspirin-intolerant asthma: a candidate gene approach. Hum Mol Genet. 2004; 13:3203–3217. PMID: 15496426.

27. Szczeklik W, Sanak M, Szczeklik A. Functional effects and gender association of COX-2 gene polymorphism G-765C in bronchial asthma. J Allergy Clin Immunol. 2004; 114:248–253. PMID: 15316498.

28. Kim SH, Choi JH, Park HS, Holloway JW, Lee SK, Park CS, et al. Association of thromboxane A2 receptor gene polymorphism with the phenotype of acetyl salicylic acid-intolerant asthma. Clin Exp Allergy. 2005; 35:585–590. PMID: 15898979.

29. Tornhamre S, Ehnhage A, Kolbeck KG, Edenius C, Lindgren JA. Uncoupled regulation of leukotriene C4 synthase in platelets from aspirin-intolerant asthmatics and healthy volunteers after aspirin treatment. Clin Exp Allergy. 2002; 32:1566–1573. PMID: 12569976.
30. Kim SH, Bae JS, Oh JM, Suh CH, Nahm DH, Park HS. Genetic polymorphism of FcεR1β-109T > C is associated with a susceptibility to aspirin-intolerant asthma. J Asthma Allergy Immunol. 2005; 25:S248.
31. Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000; 100:655–669. PMID: 10761931.

32. Akahoshi M, Obara K, Hirota T, Matsuda A, Hasegawa K, Takahashi N, et al. Functional promoter polymorphism in the TBX21 gene associated with aspirin-induced asthma. Hum Genet. 2005; 117:16–26. PMID: 15806396.

33. Kim SH, Choi JH, Lee KW, Kim SH, Shin ES, Oh HB, et al. The HLA-DRB1*1302-DQB1*0609-DPB1*0201 haplotype may be a strong genetic marker for aspirin-induced urticaria. Clin Exp Allergy. 2005; 35:339–344. PMID: 15784113.
34. Mastalerz L, Setkowicz M, Sanak M, Szczeklik A. Hypersensitivity to aspirin: common eicosanoid alterations in urticaria and asthma. J Allergy Clin Immunol. 2004; 113:771–775. PMID: 15100686.

35. Torres-Galvan MJ, Ortega N, Sanchez-Garcia F, Blanco C, Carrillo T, Quiralte J. LTC4-synthase A-444C polymorphism: lack of association with NSAID-induced isolated periorbital angioedema in a Spanish population. Ann Allergy Asthma Immunol. 2001; 87:506–510. PMID: 11770699.
36. Kim SH, Choi JH, Holloway JW, Suh CH, Nahm DH, Park CS, et al. Leukotriene-related gene polymorphisms in patients with ASA-induced urticaria and ASA-intolerant asthma: differing contributions of ALOX5 polymorphism in Korean population. J Korean Med Sci. 2005; (in press).
37. Choi JH, Kim SH, Suh CH, Nahm DH, Park HS. Polymorphisms of high-affinity IgE receptor and histamine-related genes in patients with ASA-induced urticaria/angioedema. J Korean Med Sci. 2005; 20:367–372. PMID: 15953854.

38. Bae JS, Kim SH, Kang YM, Suh CH, Nahm DH, Park HS. Significant association of FcεR1α promoter polymorphism with aspirin-intolerant chronic urticaria. J Asthma Allergy Immunol. 2005; 25:S249.
39. Kowalski ML, Ptasinska A, Jedrzejczak M, Bienkiewicz B, Cieslak M, Grzegorczyk J, et al. Aspirin-triggered 15-HETE generation in peripheral blood leukocytes is a specific and sensitive aspirin-sensitive patients identification test (ASPITest). Allergy. 2005; 60:1139–1145. PMID: 16076298.

41. Dahlen SE, Malmstrom K, Nizankowska E, Dahlen B, Kuna P, Kowalski M, et al. Improvement of aspirin-intolerant asthma by montelukast, a leukotriene antagonist: a randomized, double-blind, placebo-controlled trial. Am J Respir Crit Care Med. 2002; 165:9–14. PMID: 11779723.
42. Obase Y, Shimoda T, Tomari SY, Mitsuta K, Kawano T, Matsuse H, et al. Effects of pranlukast on chemical mediators in induced sputum on provocation tests in atopic and aspirin-intolerant asthmatic patients. Chest. 2002; 121:143–150. PMID: 11796443.

43. Micheletto C, Tognella S, Visconti M, Pomari C, Trevisan F, Dal Negro RW. Montelukast 10 mg improves nasal function and nasal response to aspirin in ASA-sensitive asthmatics: a controlled study vs placebo. Allergy. 2004; 59:289–294. PMID: 14982510.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download