Abstract
Polymorphisms of DNA repair genes, X-ray repair cross-complementing group 1 (XRCC1) might contribute to individual susceptibility to different types of cancers. We analyzed the relationship between XRCC1 polymorphisms and the risk of papillary thyroid carcinoma in a Korean sample. A hospital-based case-control study was performed in 111 papillary thyroid carcinoma patients and 100 normal control subjects. XRCC1 Arg194Trp and Arg399Gln single nucleotide polymorphisms (SNPs) were genotyped by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). The XRCC1 Arg194Trp Arg/Trp genotype was significantly associated with a decreased risk of papillary thyroid carcinoma compared to that of Arg/Arg genotype (odds ratio [95% confidence intervals]; 0.550 [0.308-0.983]). There was no significant association between XRCC1 Arg399Gln genotypes and risk of papillary thyroid carcinoma. Based on these results, the XRCC1 Arg194Trp Arg/Trp genotype could be used as a useful molecular biomarker to predict genetic susceptibility for papillary thyroid carcinoma in Koreans.
Papillary thyroid carcinoma is rising worldwide and accounted for 13.1% of cancers registered in Korea in 2007 (1, 2). Etiologic risk factors for papillary thyroid cancer remain largely unknown, although childhood exposure to ionizing radiation and a history of benign thyroid nodules are known to be related to thyroid cancer (1, 3). Generally, human cancer is believed to be induced by both environmental and host factors, including individual genetic susceptibility. Genetic susceptibility is related to chromosomal aberrations or genetic polymorphisms of various genes, including those involved in DNA repair (4-7). Accumulating evidence suggests that genetic polymorphisms influence the risk of environmental carcinogenesis, and that genetic susceptibility plays an important role in the development of human cancer (8-10).
DNA repair is critical for the maintenance of genome integrity and corrects DNA damage caused by either exogenous carcinogens or endogenously-produced reactive oxygen metabolites. X-ray repair cross-complementing group 1 (XRCC1) encodes a protein that plays an important role in the base excision repair (BER) pathway for repairing single-strand DNA breaks. XRCC1 acts by interactive conjugation with poly (ADP-ribose), DNA polymerase beta, and DNA ligase III. Many studies have suggested that XRCC-lacking cells have increased sensitivity to ionizing radiation, alkylating agents, ultra-violet light and hydrogen peroxide (11-14). Numerous single-nucleotide polymorphisms (SNPs) of XRCC1 have been identified, and XRCC1 polymorphisms have been found to be associated with increased risk for various cancers including head and neck, lung, gastric, breast, esophageal, and thyroid cancers (8, 15-20).
To our knowledge, there have been only four previous reports examining XRCC1 SNPs and the risk of thyroid carcinoma (19-22), and these studies have reported conflicting data. Furthermore, up to date, a genetic study of XRCC1 SNPs in papillary thyroid cancer has not been performed in a Korean sample. In an effort to evaluate the genetic influence on risk of papillary thyroid carcinoma, we performed a genetic analysis of two known polymorphisms of the XRCC1 gene (XRCC1 Arg194Trp and XRCC1 Arg399Gln) in a Korean sample.
The hospital-based case-control study was performed from January 2004 to August 2005. The patient group consisted of 111 patients who underwent thyroidectomy and who were confirmed as having papillary thyroid carcinoma by pathologic examination. The control group consisted of 100 volunteers who visited our health promotion center for medical examinations and who were free of thyroid nodules. The control group had normal thyroid ultrasonographies and function. Subjects who had been exposed to radiation or had a prior history of cancer were excluded from the control group. The ethnicity of all subjects was Korean. The patient group consisted of 28 males and 83 females with a mean age of 46.6 yr, and the control group consisted of 57 males and 43 females with mean age of 46.7 yr (Table 1). Female was significantly higher in the case group than in the control group (P < 0.001). Peripheral blood specimens were taken from all participants and stored at -80℃ for DNA isolation.
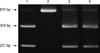
DNA was extracted from peripheral blood using the Wizard™ Genomic DNA purification kit (Promega, Madison, WI, USA). Two SNPs of XRCC1, Arg194Trp and Arg399Gln, were genotyped by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). The dimorphic sites were XRCC1 codon 194 (C→T, Arg→Trp, exon 6), and XRCC1 codon 399 (G→A, Arg→Gln, exon 10). The primers were as follows: 5'-GCC CCG TCC CAG GTA-3' and 5'-AGC CCC AAG ACC CTT TCA CT-3' to amplify a 491 bp fragment of XRCC1 covering the Arg194Trp polymorphism, and 5'-TTG TGC TTT CTC TGT GTC CA-3' and 5'-TCC TCC AGC CTT TTC TGA TA-3' to amplify a 615 bp fragment of XRCC1 covering the Arg399Gln polymorphism. Polymerase chain reaction (PCR) was performed in a mixture containing 1 × PCR buffer, 50 ng genomic DNA, 3 mM MgCl2, 200 µM dNTP mixture, 0.5 units Taq DNA polymerase, and 0.6 µM primer for codon 194 or 0.8 µM primer for codon 399. A thermal cycler (GeneAmp PCR system 2400, Perkin Elmer, MA, USA) was used with the following specifications: a denaturing step of 94℃ for 4 min, followed by 30 cycles of 94℃ for 30 sec and 68℃ for 90 sec. The PCR products were verified on 3% Metaphor agarose gels (FMC Bioproducts, Rockland, ME, USA). The restriction enzyme MspI was used to identify XRCC1 Arg194Trp and Arg399Gln SNPs. PCR products were digested with MspI restriction enzyme at 37℃ overnight, and the digested products were then separated on a 3% Metaphor agarose gels (FMC Bioproducts) containing ethidium bromide. The 491 bp fragment of codon 194 always yielded a 174 bp band, acting as an indicator of complete digestion. XRCC1 Arg194Trp genotypes Arg/Arg, Arg/Trp, and Trp/Trp generated 292 bp, 292 and 313 bp, or 313 bp DNA bands, respectively (Fig. 1). XRCC1 399Arg allele generated 221 bp and 374 bp bands upon MspI restriction, whereas XRCC1 399Gln allele generated only a 615 bp uncut band (Fig. 2).
The chi square test was used to compare XRCC1 polymorphism frequencies of the papillary thyroid carcinoma and normal control groups. A multivariate logistic regression model was used to obtain odds ratios (OR) adjusted for age and sex, with a 95% confidence interval (95% CI) for a genetic polymorphism and its association. All statistical data were obtained using SPSS (SPSS version 12.9, Chicago, IL, USA).
The genotype frequencies of the two XRCC1 SNPs are shown in Table 2. Genotype distributions of all loci were in Hardy-Weinberg equilibrium (data not shown). The frequencies of the XRCC1 Arg194Trp genotypes Arg/Arg, Arg/Trp, and Trp/Trp were 53.2%, 38.7%, and 8.1% in the cancer group and 37.0%, 49.0%, and 14.0% in the control group, respectively. The ORs (95% CI) for the XRCC1 Arg194Trp genotypes Arg/Trp and Trp/Trp relative to Arg/Arg were 0.550 (0.308-0.983) and 0.403 (0.159-1.025), respectively (Table 1). The XRCC1 Arg194Trp Arg/Trp genotype was significantly associated with a decreased risk for papillary thyroid carcinoma compared with that of the Arg/Arg genotype.
The frequencies of the XRCC1 Arg399Gln genotypes Arg/Arg, Arg/Gln and Gln/Gln were 78.4%, 15.3%, and 6.3% in the cancer group and 72.0%, 19.0%, and 9.0% in the control group, respectively. The ORs (95% CI) for XRCC1 Arg399Gln genotype Arg/Gln and Gln/Gln relative to Arg/Arg were 0.740 (0.359-1.529) and 0.644 (0.228-1.817), respectively (Table 1). The XRCC1 Arg399Gln genotypes showed a trend of decreased risk of papillary thyroid carcinoma; however, it was not significantly different from those of the other genotypes.
There were no gender- or age-associated statistically significant differences with regard to the risk of papillary thyroid carcinoma for the two XRCC1 polymorphisms (data not shown).
DNA damage is widely induced by both normal, endogenous metabolic processes and environmental carcinogens. If such damage is not repaired, it could result in mutations and genomic instability, which could in turn cause cellular malignant transformation. DNA repair mechanisms include four pathways involving more than 80 different genes with direct roles in repairing damaged DNA (10). The mismatch repair (MMR) pathway corrects primarily mispaired nucleotides arising during DNA replication. The nucleotide excision repair (NER) pathway mainly removes damage induced by ultra-violet (UV) and chemical exposure. Double-strand breaks caused by exposure to ionizing radiation or incomplete repair of other damage are repaired via double-strand-break repair (DSBR) pathways. The BER pathway is involved in the repair of damaged bases and abasic sites induced by free radicals. Many enzymes play a role in each pathway, including MLH1, MSH2, PMS2, and MSH6 in the MMR pathway; XPC, XPD, and ERCC1 in the NER pathway; BRCA1, BRCA2, and XRCC3 in the DSBR pathway; and XRCC1 in the BER pathway (10).
The XRCC1 protein functions exclusively in DNA BER, strand-break repair, and maintenance of genetic stability. Although the functional consequences of the two non-synonymous polymorphisms in XRCC1 that were analyzed in this study are not fully understood, the nature of the amino acid substitutions (Arg194Trp and Arg399Gln) may be expected to cause functional changes in the XRCC1 protein. Such functional differences could potentially affect the risk of developing cancer. However, previous studies have yielded conflicting data. For instance, the XRCC1 194Trp allele was found to be associated with an increased risk of certain cancers, but was also found to exert a protective effect against cancer (15, 16).
To our knowledge, XRCC1 SNPs and the associated risks of thyroid carcinoma were examined in four previous reports that yielded conflicting data (19-22). Chiang et al. (19) reported that the XRCC1 Arg194Trp genotype Trp/Trp has a significantly increased risk for differentiated thyroid carcinoma in Taiwanese samples (OR 1.85; 95% confidence interval, 1.11-3.07). In another study performed in the United States, Ho et al. (20) reported that the XRCC1 Arg194Trp heterozygous Arg/Trp genotype is associated with increased risk of differentiated thyroid carcinoma, while the XRCC1 Arg399Gln homozygous Gln/Gln genotype and variant Gln allele are associated with decreased risk. The risks of papillary thyroid carcinoma associated with XRCC1 Arg194Trp and Arg399Gln were also examined in a Chinese sample (21). According to this study, the XRCC1 399Gln variant genotype was associated with increased risk (OR: 2.71, 95% CI: 1.22-6.05), but the XRCC1 194Trp variant genotype was not significantly correlated. In another study performed in Kazakhstan (22), thyroid cancer risk was decreased for the minor Trp allele of XRCC1 Arg194Trp. In this study, individuals with XRCC1 Arg194Trp Arg/Trp genotype had a 0.55-fold decreased risk of papillary thyroid carcinoma compared to that of the XRCC1 Arg194Trp Arg/Arg genotype.
Although it would be hard to decipher the reasons for these contradictory results, they could be ascribed to several factors that may impact the polymorphisms in different ways, including variation in carcinogen exposures in different populations and different types of DNA damage in the initiation of different cancers. Moreover, inadequate study design such as nonrandom sampling, limited sample size, and pitfalls of unknown confounding influences should also be considered.
There are some limitations in this study. First, there was a significant difference of the distribution of gender between the cancer and control groups, although we analyzed the ORs with adjustment for gender and age. The more accurate risk estimation may be possible in gender-matched case-control samples in the future. Second, the selection bias of a hospital-based case-control study may be an issue. Specifically, the control group in our study may not have accurately represented the general population, although the genotype distribution was in Hardy-Weinberg equilibrium. Third, our study included a relatively small number of cases (n = 111) and controls (n = 100). A further study with using large, matched case-control samples is necessary to validate the genetic effects of XRCC1 polymorphisms in the Korean population.
In conclusion, this study is the first to analyze XRCC1 SNPs and their associated risk of papillary thyroid carcinoma in a Korean sample. In this study, the Arg/Trp genotype of the XRCC1 Arg194Trp polymorphism is significantly associated with a reduced risk for papillary thyroid carcinoma. The XRCC1 Arg194Trp Arg/Trp genotype could potentially be used as a molecular biomarker to predict lower risk for papillary thyroid carcinoma in Koreans.
Figures and Tables
Fig. 1
XRCC1 Arg194Trp single nucleotide polymorphism (SNP) by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). Arg/Arg genotype, lane 1 and 3; Arg/Trp genotype, lane 2 and 4; Trp/Trp genotype, lane 5.

AUTHOR SUMMARY
XRCC1 Polymorphisms and Risk of Papillary Thyroid Carcinoma in a Korean Sample
Ri A Ryu, Kyung Tae, Hyun Jung Min, Jin Hyeok Jeong, Seok Hyun Cho, Seung Hwan Lee and You Hern Ahn
Polymorphisms of DNA repair genes, X-ray repair cross-complementing group 1 (XRCC1) might contribute to individual susceptibility to papillary thyroid carcinoma in the Korean population. We genotyped XRCC1 Arg194Trp and Arg399Gln single nucleotide polymorphisms (SNPs) in 111 papillary thyroid carcinoma patients and 100 normal control subjects. The XRCC1 Arg194Trp Arg/Trp genotype was significantly associated with a decreased risk of papillary thyroid carcinoma compared to that of Arg/Arg genotype. Based on these results, the XRCC1 Arg194Trp Arg/Trp genotype could be used as a useful molecular biomarker to predict genetic susceptibility for papillary thyroid carcinoma in the Korean.
References
1. How J, Tabah R. Explaining the increasing incidence of differentiated thyroid cancer. CMAJ. 2007. 177:1383–1384.
2. Jung KW, Park S, Kong HJ, Won YJ, Boo YK, Shin HR, Park EC, Lee JS. Cancer statistics in Korea: incidence, mortality and survival in 2006-2007. J Korean Med Sci. 2010. 25:1113–1121.
3. Shore RE. Issues and epidemiological evidence regarding radiation-induced thyroid cancer. Radiat Res. 1992. 131:98–111.
4. Spitz MR, Fueger JJ, Beddingfield NA, Annegers JF, Hsu TC, Newell GR, Schantz SP. Chromosome sensitivity to bleomycin-induced mutagenesis, an independent risk factor for upper aerodigestive tract cancers. Cancer Res. 1989. 49:4626–4628.
5. Wang LE, Sturgis EM, Eicher SA, Spitz MR, Hong WK, Wei Q. Mutagen sensitivity to benzo(a)pyrene diol epoxide and the risk of squamous cell carcinoma of the head and neck. Clin Cancer Res. 1998. 4:1773–1778.
6. Cheng L, Eicher SA, Guo Z, Hong WK, Spitz MR, Wei Q. Reduced DNA repair capacity in head and neck cancer patients. Cancer Epidemiol Biomarkers Prev. 1998. 7:465–468.
7. Schantz SP, Hsu TC, Ainslie N, Moser RP. Young adults with head and neck cancer express increased susceptibility to mutagen-induced chromosome damage. JAMA. 1989. 262:3313–3315.
8. Tae K, Lee HS, Park BJ, Park CW, Kim KR, Cho HY, Kim LH, Park BL, Shin HD. Association of DNA repair gene XRCC1 polymorphisms with head and neck cancer in Korean population. Int J Cancer. 2004. 111:805–808.
9. Ji YB, Tae K, Lee YS, Lee SH, Kim KR, Park CW, Park BL, Shin HD. XPD polymorphisms and risk of squamous cell carcinoma of the head and neck in a Korean sample. Clin Exp Otorhinolaryngol. 2010. 3:42–47.
10. Hu JJ, Mohrenweiser HW, Bell DA, Leadon SA, Miller MS. Symposium overview: genetic polymorphisms in DNA repair and cancer risk. Toxicol Appl Pharmacol. 2002. 185:64–73.
11. Kubota Y, Nash RA, Klungland A, Schär P, Barnes DE, Lindahl T. Reconstruction of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996. 15:6662–6670.
12. Izumi T, Hazra TK, Boldogh I, Tomkinson AE, Park MS, Ikeda S, Mitra S. Requirement for human AP endonuclease 1 for repair of 3'-blocking damage at DNA single-strand breaks induced by reactive oxygen species. Carcinogenesis. 2000. 21:1329–1334.
13. Zhang X, Moréra S, Bates PA, Whitehead PC, Coffer AI, Hainbucher K, Nash RA, Sternberg MJ, Lindahl T, Freemont PS. Structure of an XRCC1 BRCT domain: a new protein-protein interaction module. EMBO J. 1998. 17:6404–6411.
14. Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interaction. EMBO J. 2001. 20:6530–6539.
15. Patel AV, Calle EE, Pavluck AL, Feigelson HS, Thun MJ, Rodriguez C. A prospective study of XRCC1 (X-ray cross-complementing group 1) polymorphisms and breast cancer risk. Breast Cancer Res. 2005. 7:R1168–R1173.
16. Wang Y, Yang H, Li H, Li L, Wang H, Liu C, Zheng Y. Association between X-ray repair cross complementing group 1 codon 399 and 194 polymorphisms and lung cancer risk: a meta-analysis. Cancer Lett. 2009. 285:134–140.
17. Dai L, Wang K, Zhang J, Lv Q, Wu X, Wang Y. XRCC1 gene polymorphisms and esophageal squamous cell carcinoma risk in Chinese population: a meta-analysis of case-control studies. Int J Cancer. 2009. 125:1102–1109.
18. Shen H, Xu Y, Qian Y, Yu R, Qin Y, Zhou L, Wang X, Spitz MR, Wei Q. Polymorphisms of the DNA repair gene XRCC1 and risk of gastric cancer in a Chinese population. Int J Cancer. 2000. 88:601–606.
19. Chiang FY, Wu CW, Hsiao PJ, Kuo WR, Lee KW, Lin JC, Liao YC, Juo SH. Association between polymorphism in DNA base excision repair genes XRCC1, APE1, and ADPRT and differentiated thyroid carcinoma. Clin Cancer Res. 2008. 14:5919–5924.
20. Ho T, Li G, Lu J, Zhao C, Wei Q, Sturgis EM. Association of XRCC1 polymorphisms and risk of differentiated thyroid carcinoma: a case-control analysis. Thyroid. 2009. 19:129–135.
21. Zhu QX, Bian JC, Shen Q, Jiang F, Tang HW, Zhang HW, Wu Y. Genetic polymorphisms in X-ray repair cross-complementing gene 1 and susceptibility to papillary thyroid carcinoma. Zhonghua Liu Xing Bing Xue Za Zhi. 2004. 25:702–705.
22. Sigurdson AJ, Land CE, Bhatti P, Pineda M, Brenner A, Carr Z, Gusev BI, Zhumadilov Z, Simon SL, Bouville A, Rutter JL, Ron E, Struewing JP. Thyroid nodule, polymorphic variants in DNA repair and RET-related genes, and interaction with ionizing radiation exposure from nuclear tests in Kazakhstan. Radiat Res. 2009. 171:77–88.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download