Abstract
Transforming growth factor-β1 (TGF-β1) is a potent inhibitor of cellular growth and proliferation by G1 phase arrest or apoptosis. We investigated the association of TGF-β1 with the anti-proliferative effect of upstream stimulatory factor (USF) in Fischer rat thyroid cell line (FRTL-5) cells. [Methyl-3H] thymidine uptake was measured after treatment of FRTL-5 cells with TGF-β1 to identify its anti-proliferative effect. USF-1 and USF-2 proteins were in vitro translated, and an electrophoretic mobility shift assay was performed to identify the interaction between USF and the TGF-β1 promoter. FRTL-5 cells were transfected with USF cDNA, and then the expression of TGF-β1 was examined with Northern and Western blotting. The cell cycle-regulating proteins associated with TGF-β1 were also measured. TGF-β1 significantly inhibited [methyl-3H] thymidine uptake in FRTL-5 cells. Two specific binding sites for USF were found in the TGF-β1 promoter: -1,846~-1,841 (CACATG) and -621~-616 (CATGTG). Overexpression of USF increased both the mRNA levels and protein levels of TGF-β1. However, the expression of cyclin D1, CDK4, cyclin E, and CDK2, and the phosphorylation of retinoblastoma protein remained unchanged. Overexpression of USF in FRTL-5 cells increased the expression of TGF-β10 through specific binding to TGF-β1 promoter. However, the USF-induced expression of TGF-β1 did not cause G1 arrest.
Upstream stimulatory factor (USF) is a ubiquitous transcription factor in mammalian cells. It was originally identified as a cellular transcription factor, which is important for expression of the adenovirus major late promoter (1, 2). Two different genes, defined as USF-1 and USF-2, have been isolated from HeLa cells, and these genes encode 43 and 44 kDa polypeptides, respectively (3-6). These two peptides have identical DNA-binding specificities and transcriptional activities (3-6). USF-1 and USF-2 polypeptides are highly divergent in their N-terminal sequences, but they share a highly conserved C-terminal basic/helix-loop-helix/leucine zipper (B-HLH-LZ) sequence that is important for the dimerization and DNA-binding of USF (6). Proteins of the B-HLH-LZ gene family bind to the DNA sequences of the general type CANNTG, which is referred to as the E-box (7, 8). USF has been reported to inhibit cellular growth and proliferation in some cell types (9, 10). We have recently observed that USF also inhibits the cellular proliferation of rat thyroid cells, Fischer rat thyroid cell line (FRTL-5) cells, and papillary thyroid carcinoma cells (11).
Transforming growth factor-β1 (TGF-β1) is a potent, naturally occurring inhibitor of cellular growth and proliferation (12, 13). The role of TGF-β in tumorigenesis can be suppressive or promoting depending on the developmental stage of a tumor and its cellular context (14). During an early phase of tumorigenesis, TGF-β inhibits tumor development and growth by inducing the arrest of the cell cycle and possibly apoptosis (14). TGF-β1 is known to bind to FRTL-5 cells and inhibit their cellular growth and proliferation (15-18).
Because both TGF-β1 and USF suppress FRTL-5 cell proliferation, and because USF increases the activity of TGF-β1 in some cell types (19), we investigated the effect of USF on the expression and function of TGF-β1 in order to clarify the mechanism of the anti-proliferative effect of USF in FRTL-5 cells. First of all, we assessed whether USF binds to the TGF-β1 promoter. Second, we investigated the effect of USF on the expression of TGF-β1 and the related cell cycle regulatory proteins in FRTL-5 cells.
Fresh F1 subclone of FRTL-5 cells (Interthyr Research Foundation, Baltimore, MD, U.S.A.) was grown in a 37℃, 5% CO2-95% O2 incubator with 6H medium that consisted of Coon's modified Ham's F-12 (Sigma Chemical, St. Louis, MO, U.S.A.) supplemented with 5% calf serum, 1 mM nonessential amino acids, 2 mM glutamine, and a mixture of 6 hormones: bovine thyroid stimulating hormone (TSH) (10 mU/mL), insulin (10 µg/mL), hydrocortisone (0.4 ng/mL), transferrin (5 µg/mL), glycyl-L-histidyl-L-lysine acetate (2-10 ng/mL), and somatostatin (10 ng/mL). The culture media were replaced with fresh medium every 3 days, and the cells were passaged every 7-10 days. In some experiments, the cells were maintained in 5H medium without TSH for an additional 5-6 days after 70-80% cell confluence was achieved in the 6H medium.
FRTL-5 cells were maintained in 5H medium after being grown in 6H medium, and they were incubated with 5 ng/mL of TGF-β1 (Calbiochem of EMD Biosciences, Inc, Darmstadt, Germany) in a 37℃, 5% CO2 humidified incubator for 36 hr.
Two or three uCi/well of [methyl-3H] thymidine (Perkin-Elmer, Wellesley, MA, U.S.A.) was added, and then the cells were incubated in an incubator maintained at 37℃ for an additional 6 hr. The cells were washed twice with cold phosphate buffered saline (PBS), and were then incubated with 1 mL of 5% trichloroacetic acid for 20 min at room temperature. The cells were incubated again with 1 mL of 0.5 N KOH overnight after being washed with 5% trichloroacetic acid and PBS. [methyl-3H] thymidine uptake was measured using a multi-purpose scintillation counter (Beckman Coulter Inc., Fullerton, CA, U.S.A.), and this was expressed as counts per minute (cpm).
Total RNA was extracted from the FRTL-5 cells using an RNA extraction kit (Invitrogen, Carlsbad, CA, U.S.A.), and the amount of total RNA was measured using a spectrophotometer (Pharmacia, Buckinghamshire, England). Two micrograms of extracted RNA mixed with 0.1% diethyl pyrocarbonate (DEPC)-treated distilled water were incubated for 5 min at 70℃. The cDNA was synthesized by incubation for 60 min at 42℃ using a commercial kit (AccuPower RT PreMix, BiONEER, Daejeon, Korea). Polymerase chain reaction (PCR) was performed in a 20 µL reaction mixture containing 5 µL of denatured cDNA, 250 µM of dNTP mix, 2.5 U of Taq DNA polymerase (BiONEER), 20 mM of Tris-HCl (pH 8.8), 10 mM of KCl, 2 mM of MgSO4, and 0.5 µM of each primer (the primer sequences are listed in Table 1). PCR was carried out in a thermal cycler Gene-Amp 9600 PCR apparatus (Perkin-Elmer Corp., Norwalk, CT, U.S.A.) according to the following protocol: initial denaturation at 95℃ for 5 min, followed by 30 cycles of denaturation at 94 to 95℃ for 30 sec to 1 min, annealing at 52 to 60℃ for 30 sec to 1 min, and extension at 72℃ for 1 to 2 min, and completed by a final extension step at 72℃ for 10 min. Ten microliters of PCR product were analyzed by electrophoresis in 1.5% agarose gel. The PCR products were purified using a purification kit (PCRquick-spin PCR Product Purification Kit, iNtRON Biotechnology, Seongnam, Korea), and were then cloned into the pcDNA3.1 vectors using a pcDNA3.1/V5-His TOPO TA expression kit (Invitrogen). The cDNA was extracted from a cloned vector using a QIAprep Spin Miniprep kit (QIAGEN Inc, Valencia, CA, U.S.A.). Gene sequencing was performed using an automated direct sequencing analyzer (ABI PRISM, 377 DNA sequencer, Perkin Elmer, Foster City, CA, U.S.A.).
In vitro translated proteins were made from USF-1 and USF-2 cDNAs that were cloned into the pcDNA3.1 vectors using a TNT T7 Quick Coupled Transcription/Translation System (Promega, Madison, WI, U.S.A.). The amount of synthetic protein was calculated using a commercial kit assay (Bio-Rad, Hercules, CA, U.S.A.), and was confirmed by Western blotting and then used for the electrophoretic mobility shift assay (EMSA).
Four types of E-boxes exist in the rat TGF-β1 promoter (E1-E4, Table 2). The sense and antisense strand oligonucleotides that contained each of the E-box sequences were annealed, and the 5' end was labeled with [γ-32P]ATP by using T4 polynucleotide kinase (Promega). The upper-strand sequences of the double-stranded oligonucleotides were used as probes (T1-T4), and they are shown in Table 2.
The EMSA reaction mixtures included 10 µg of synthetic protein and reaction buffer (10 mM HEPES; pH 7.9, 60 mM NaCl, 1 mM EDTA, 2 mM dithiothreitol, 1 µg poly (dIdC) and 7% glycerol). After incubation for 10 min at room temperature, 300,000 cpm of radiolabeled probe was added, and the reaction was allowed to proceed for an additional 45 min. The reaction mixture was electrophoresed on a 6% polyacrylamide gel at 300 V for 2 hr using tris-borate-EDTA buffer. The gel was dried and exposed to radiography film overnight at -70℃. Specificity of binding was ascertained by competition with a 160-fold molar excess of unlabeled oligonucleotides and by conducting supershift experiments. For the supershift assay, 2 µL (0.2 µg/µL) of anti-USF-1 or anti-USF-2 antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, U.S.A.) was preincubated for 2 hr on ice before the addition of the probes.
FuGENE6 (Roche Diagnostics, Mannheim, Germany) was used to transfect DNA plasmids into FRTL-5 cells. FRTL-5 cells were plated onto 6 cm culture dishes, and were grown to 50-60% confluence in 6H medium and then transferred to 0.2% calf serum with 4H medium (excluding TSH and insulin) for 3 days. Eighteen hours prior to transfection, the medium was changed to 6H medium. Cells were then cotransfected with the pcDNA3 vector (5 µg) or the USF-expression vector (5 µg) plus an expression vector that coded for green fluorescent protein (5 µg, pEGFP; Clontech, Sydney, Australia). The cells were harvested 30 or 50 hr after transfection with FuGene6. The average transfection efficiency into the FRTL-5 cells was 15% to 30%.
Total RNA was isolated from FRTL-5 cells that were transfected with an empty vector or the USF-expression vector using a commercial kit (QIAGEN Inc). Five micrograms of total RNA were run on a denatured 1% agarose gel (final formaldehyde concentration: 2.2 M); they were transferred to a positively-charged nylon membrane (Roche) and immobilized by UV cross-linking. Full-length rat USF-1, mouse USF-2, rat TGF-β1, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNAs were used as probes and were synthesized using a commercial kit (PCR DIG Probe Synthesis Kit, Roche). Hybridization was carried out overnight at 50℃ with 50 ng of cDNA probe in DIG Easy Hyb (Roche). Subsequent washing and detection with CSPD (Roche) were carried out as recommended by the protocol of the manufacturer. Each experiment was conducted twice. The degree of RNA expression was expressed as values that were adjusted by those of GAPDH.
To obtain the total cellular proteins, FRTL-5 cells transfected with empty PCDNA3.1 vector or with the USF-expression vector were resuspended in RIPA buffer (50 mM tris-Cl; pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid and 0.1% SDS) supplemented with NaF, Na3VO4, phenylmethylsulfonyl fluoride (PMSF), and complete protease inhibitor cocktail tablets (Roche) after the cells had been washed with cold PBS. The cell suspension was incubated on ice for 10 min and then centrifuged at 15,000×g for 20 min at 4℃. The resultant supernatant contained the total cellular protein. The amount of cellular protein was calculated using a commercial kit assay (Bio-Rad, Hercules, CA, U.S.A.). For the Western immunoblots, 20 µg of protein was analyzed on SDS-PAGE, transferred to nitrocellulose membranes (Amersham, Germany), blocked in 5% skim milk, incubated with primary antibodies overnight at 4℃, and then revealed by treatment with SuperSignal®. West Femto Maximum Sensitivity Substrate (Pierce, Rockford, IL, U.S.A.). Antibody to retinoblastoma protein (Rb) used for Western blot analysis was purchased from BD Pharmingen (San Diego, CA, U.S.A.), and the other antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, U.S.A.).
All of the experiments were repeated at least three times with different batches of cells. The data for thymidine uptake of FRTL-5 cells were analyzed using the SPSS-PC software program (version 11.0, Chicago, IL, U.S.A.). The values of thymidine uptake were analyzed using the Student's t-test, and were expressed as mean±standard deviation. p values less than 0.05 were considered significant.
Treatment with 5 ng/mL of TGF-β1 significantly decreased [methyl-3H]-thymidine uptake by 60-70% in FRTL-5 cells (Fig. 1).
USF-1 and USF-2 proteins were in vitro translated and then confirmed by Western blotting (right upper box of Fig. 2). To identify binding of USF-1 and USF-2 proteins to the TGF-β1 promoter, we observed the DNA-protein complex between four different E-box sequences of the rat TGF-β1 promoter (T1, T2, T3, and T4 in Table 2) and the USF proteins by performing EMSA (as indicated by the black arrows in Fig. 2). Among them, the affinities for binding to T2 and T3 were much stronger. Cold competitors eliminated the DNA-protein complexes, and a specific anti-USF-1 or anti-USF-2 antibody shifted these complexes to an upper position to make 'super-shifts' (as indicated by the white arrows in Fig. 2). Two loci of E2 (5'-CACATG) and E3 (5'-CATGTG) were detected in the rat TGF-β1 promoter, respectively: -1,846~-1,841 (E2), -3,514~-3,509 (E2'), and -621~ -616 (E3), -1,563~-1,558 (E3'). The specific sites for USF binding were evaluated by performing EMSA with four probes (T2, T2', T3, and T3') that contained each E-box, and were identified as -1,846~-1,841 (E2) and -621~-616 (E3) (Fig. 3).
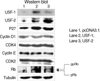
Transient transfection with USF-1 and USF-2 induced their overexpression in FRTL-5 cells. Overexpression of USF-1 and USF-2 increased both the mRNA levels and protein levels of TGF-β1 in FRTL-5 cells (Fig. 4).
The expression of G1 phase-related proteins, including cyclin D1, CDK4, cyclin E, and CDK2, remained unchanged by USF overexpression, although the levels of p27, a cell cycle inhibitor, were increased. In addition, the overexpression of USF-1 and USF-2 did not alter the levels of phosphorylation of Rb protein in FRTL-5 cells (Fig. 5).
USF has been reported to inhibit cellular growth and proliferation in some cell types (9, 10). We have recently observed that both USF-1 and USF-2 play a role in suppressing the cellular proliferation of normal thyrocytes and thyroid papillary carcinoma cells, while they maintained the TSH-stimulated production of cAMP (11). However, the exact mechanism of the anti-proliferative effect of USF overexpression was not identified. TGF-β1 is a potent, naturally occurring inhibitor of cell growth (13), it has a high affinity for FRTL-5 cells, and it inhibits their cellular growth (15-18); therefore, this study examined the role of TGF-β1 in FRTL-5 cells overexpressing USF.
Inhibition of cellular growth by TGF-β1 has been observed in normal feline thyroid cells, human thyroid cells, and the human papillary thyroid carcinoma cell line (17, 20). First of all, we confirmed that TGF-β1 significantly decreased the uptake of [methyl-3H]-thymidine in FRTL-5 cells. Therefore, we conducted a study on the relationship between USF and TGF-β1. There have been a few reports on this relationship thus far: USF-1 and USF-2 are known to bind to the TGF-β1 promoter and increase its activity in mesangial cells, glioblastoma, embryonal carcinoma, breast carcinoma, and choriocarcinoma cells (19, 21, 22). However, the effect of USF on the expression of TGF-β1 in thyrocytes has not yet been reported. USF binds to DNA as a homodimer or heterodimer at a specific site that is characterized by an E-box (5'-CANNTG) (3, 5, 9). In this study, we demonstrated that USF-1 and USF-2 proteins strongly bound to the specific sequences (CACATG in -1,846 to -1,841 and CATGTG in -621 to -616) of the TGF-β1 promoter, and the overexpression of USF increased the mRNA and protein levels of TGF-β1 in FRTL-5 cells. Other E-boxes such as E1 and E4 in Fig. 2 were very week in their affinity to USF, which suggests that there are other factors than nucleotide sequences determining the binding of USF at E-box. It is apparent in the Fig. 3 because even E-boxes having the same sequences in the TGF-β1 promoter showed different affinity to USF. Taking into consideration the anti-proliferative effect of TGF-β1, these findings suggested that USF might inhibit cell proliferation through the induction of TGF-β1 after binding to its promoter.
As commented on earlier, TGF-β regulates cellular growth and proliferation in a cell-specific manner. In most epithelial cells, TGF-β is a potent inhibitor of cellular growth and proliferation; it arrests the cell cycle in the G1 phase by modulating the cell cycle regulators, especially the CDK inhibitors, CDK4, CDK2, cyclin D, and cyclin E, and by eventually dephosphorylating Rb protein (23). TGF-β induces the expression of CDK inhibitors such as p15, p21, and p27 (24). They, in turn, downregulate CDK4 and G1/S phosphatase CDC25A (25, 26), and they decrease the expression of c-Myc, which again down-regulates p15 (27). Inactivation of the CDK-cyclin complexes contributes to dephosphorylation of Rb protein (28). Rb protein is a primary target of the action of TGF-β1, and as Rb dephosphorylation induces G1 phase arrest (29), the main anti-proliferative mechanism of TGF-β1 on the thyroid cells has been reported as G1 phase arrest (16, 18). However, we were not able to observe G1 arrest in the USF-overexpressing FRTL-5 cells, although the expression of TGF-β1 was increased. We did not evaluate the mechanism for it, but overexpression of USF might influence the activity of G1 phase-related cyclin modulators and the phosphorylation status of Rb protein. Instead, we observed G2/M phase arrest (11). The G2/M phase arrest was accompanied by the modification of the expression of the G2/M-related regulators such as cyclin B1 and CDK1 (11). The effect of TGF-β1 on the progression of the G2/M phase is not yet clear. We observed the increase of p27 which could contribute to the G2/M phase arrest. Hashimoto et al. recently reported that TGF-β1 induced the cell cycle arrest not at G1 phase but at G2 phase in some hepatoma cells, which was thought to be through the stabilization of Wee1 (30). Although we did not examine the activity of Wee1, the overexpression of USF might affect Wee1 activity and induce G2/M arrest. However, we cannot exclude the possibility that the role of TGF-β1 in the USF-regulated anti-proliferative effects on FRTL-5 cells is very limited, regardless of the binding activities of USFs to TGF-β1 promoter.
In conclusion, we demonstrated that TGF-β1 decreased the proliferation of FRTL-5 cells. We also observed that overexpression of USF-1 and USF-2 increased the expression of TGF-β1 in FRTL-5 cells after specifically binding to the TGF-β1 promoter. However, dephosphorylation of Rb was not induced, and this was concordant with the finding that cell cycle arrest was observed at the G2/M phase, and was not seen at the G1 phase (11). The mechanism related to the reason that USF-induced TGF-β1 expression did not cause dephoshorylation of Rb protein will be evaluated and reported on in the future.
Figures and Tables
Fig. 1
Inhibition of cellular proliferation by TGF-β1 in FRTL-5 cells. FRTL-5 cells were cultured with 5 ng/mL of TGF-β1, and thymidine uptake of the cells was measured. The values of thymidine uptake were analyzed using Student's t-test, and were expressed as mean±standard deviation. Treatment with TGF-β1 significantly decreased thymidine uptake by 60-70% in FRTL-5 cells.
*p<0.05 vs. no TGF-β1 treatment.
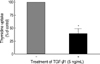
Fig. 2
Electrophoretic mobility shift assay (EMSA) of the complexes between the USF protein and the E-box sequences in the TGF-β1 promoter. The in vitro translated USF-1 and USF-2 were confirmed by Western blotting, and this is illustrated in a box located in the right upper side of the figure. Four types of E-boxes (E1-E4) existed in the TGF-β1 promoter, and each was tested to determine whether they would bind to USF-1 and USF-2 by using EMSA (black arrows). Among them, the affinities to E2 and E3 were much stronger than the affinities shown to E1 and E4. Cold competitors eliminated the DNA-protein complexes, and a specific anti-USF-1 or anti-USF-2 antibody shifted these complexes to an upper position to cause 'super-shifts' (white arrows).

Fig. 3
Electrophoretic mobility shift assay (EMSA) of the complexes between USF and the E2/E3 of the different loci in the TGF-β1 promoter. There were two loci of CACATG (E2) and CATGTG (E3), respectively: -1,846~-1,841 (E2) and -3,514~-3,509 (E2'), and -621~-616 (E3) and -1,563~-1,558 (E3'). Specific sites for USF binding were identified by performing EMSA with four probes (T2, T2', T3, and T3') that contained each E-box. The specific sites were revealed as -1,846~-1,841 and -621~-616. Black arrows indicate the DNA-protein complexes, and the white arrows indicate their super-shifts according to the specific antibodies.

Fig. 4
Effects of USF-1 and USF-2 overexpression on the expression of TGF-β1 in the FRTL-5 cells. Total RNA and protein were isolated from FRTL-5 cells that were transiently overexpressed with USF. Northern (left panel) and Western blotting (right panel) were performed to evaluate the mRNA and protein levels of USF and TGF-β1. Expression of TGF-β1 was induced by the overexpression of USF-1 and USF-2. Three sets of experiments were repeated. GAPDH was used as the internal control for Northern blotting, and tubulin was used as the internal control for Western blotting.

Fig. 5
Effects of USF-1 and USF-2 overexpression on the expression of TGF-β1-related proteins in FRTL-5 cells. Total protein was isolated from FRTL-5 cells that transiently overexpressed USF, and Western blotting was then performed. The p27 levels were increased by USF overexpression, but the cyclin D1, CDK4, cyclin E, and CDK2 levels were not changed. Accordingly, dephosphorylation of retinoblastoma (Rb) protein was not observed. Three sets of experiments were performed, and tubulin was used as the internal control.
ppRb, hyperphosphorylated Rb; pRb, hypophosphorylated Rb.
References
1. Sawadogo M, Roeder RG. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985. 43:165–175.

2. Carthew RW, Chodosh LA, Sharp PA. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell. 1985. 43:439–448.

3. Sawadogo M, Van Dyke MW, Gregor PD, Roeder RG. Multiple forms of the human gene-specific transcription factor USF. I. Complete purification and identification of USF from HeLa cell nuclei. J Biol Chem. 1988. 263:11985–11993.

4. Sawadogo M. Multiple forms of the human gene-specific transcription factor USF. II. DNA binding properties and transcriptional activity of the purified HeLa USF. J Biol Chem. 1988. 263:11994–12001.

5. Gregor PD, Sawadogo M, Roeder RG. The adenovirus major late transcription factor USF is a member of the helix-loop-helix group of regulatory proteins and binds to DNA as a dimmer. Genes Dev. 1990. 4:1730–1740.
6. Sirito M, Walker S, Lin Q, Kozlowski MT, Klein WH, Sawadogo M. Members of the USF family of helix-loop-helix proteins bind DNA as homo- as well as heterodimers. Gene Expr. 1992. 2:231–240.
7. Miyamoto NG, Moncollin V, Wintzerith M, Hen R, Egly JM, Chambon P. Stimulation of in vitro transcription by the upstream element of the adenovirus-2 major late promoter involves a specific factor. Nucleic Acids Res. 1984. 12:8779–8799.
8. Yu YT, Manley JL. Generation and functional analyses for base-substitution mutants of the adenovirus 2 major late promoter. Nucleic Acids Res. 1984. 12:9309–9321.

9. Luo X, Sawadogo M. Antiproliferative properties of the USF family of helix-loop-helix transcription factors. Proc Natl Acad Sci USA. 1996. 93:1308–1313.

10. Qyang Y, Luo X, Lu T, Ismail PM, Krylov D, Vinson C, Sawadogo M. Cell-type-dependent activity of the ubiquitous transcription factor USF in cellular proliferation and transcriptional activation. Mol Cell Biol. 1999. 19:1508–1517.

11. Jung HS, Kim KS, Chung YJ, Chung HK, Min YK, Lee MS, Lee MK, Kim KW, Chung JH. USF inhibits cell proliferation through delay in G2/M phase in FRTL-5 cells. Endocr J. 2007. 54:275–285.

12. Glick AB. TGF beta1 back to the future: revisiting its role as a transforming growth factor. Cancer Biol Ther. 2004. 3:276–283.
13. Kaklamani VG, Pasche B. Role of TGF-beta in cancer and the potential for therapy and prevention. Expert Rev Anticancer Ther. 2004. 4:649–661.
14. Sun L. Tumor-suppressive and promoting function of transforming growth factor beta. Front Biosci. 2004. 9:1925–1935.

15. Pang XP, Park M, Hershman JM. Transforming growth factor-beta blocks protein kinase-A-mediated iodide transport and protein kinase-C-mediated DNA synthesis in FRTL-5 rat thyroid cells. Endocrinology. 1992. 131:45–50.

16. Coppa A, Mincione G, Mammarella S, Ranieri A, Colletta G. Epithelial rat thyroid cell clones, escaping from transforming growth factor beta negative growth control, are still inhibited by this factor in the ability to trap iodide. Cell Growth Differ. 1995. 6:281–290.
17. Asmis LM, Kaempf J, Von Gruenigen C, Kimura ET, Wagner HE, Studer H. Acquired and naturally occurring resistance of thyroid follicular cells to the growth inhibitory action of transforming growth factor-beta 1 (TGF-beta 1). J Endocrinol. 1996. 149:485–496.
18. Carneiro C, Alvarez CV, Zalvide J, Vidal A, Dominguez F. TGF-beta1 actions on FRTL-5 cells provide a model for the physiological regulation of thyroid growth. Oncogene. 1998. 16:1455–1465.
19. Weigert C, Brodbeck K, Sawadogo M, Haring HU, Schleicher ED. Upstream stimulatory factor (USF) proteins induce human TGF-beta1 gene activation via the glucose-response element-1013/-1002 in mesangial cells: up-regulation of USF activity by the hexosamine biosynthetic pathway. J Biol Chem. 2004. 279:15908–15915.
20. Pang XP, Hershman JM. Transforming growth factor-beta1 resistance in a thyroid cancer model of tumor necrosis factor-alpha resistance. Thyroid. 1998. 8:1065–1070.
21. Scholtz B, Kingsley-Kallesen M, Rizzino A. Transcription of the transforming growth factor-beta2 gene is dependent on an E-box located between an essential cAMP response element/activating transcription factor motif and the TATA box of the gene. J Biol Chem. 1996. 271:32375–32380.
22. Kingsley-Kallesen M, Luster TA, Rizzino A. Transcriptional regulation of the transforming growth factor-beta2 gene in glioblastoma cells. In Vitro Cell Dev Biol Anim. 2001. 37:684–690.
23. Ravitz MJ, Wenner CE. Cyclin-dependent kinase regulation during G1 phase and cell cycle regulation by TGF-beta. Adv Cancer Res. 1997. 71:165–207.
24. Massague J, Blain SW, Lo RS. TGF beta signaling in growth control, cancer, and heritable disorders. Cell. 2000. 103:295–309.
25. Ewen ME, Sloss HK, Whitehouse LL, Livingston DM. TGF beta inhibition of CDK4 synthesis is linked to cell cycle arrest. Cell. 1993. 74:1009–1020.
26. Iavarone A, Massague J. Repression of the CDK activator cdc25A and cell-cycle arrest by cytokine TGF-beta in cells lacking the CDK inhibitor p15. Nature. 1997. 387:417–422.
27. Warner BJ, Blain SW, Seoane J, Massague J. Myc downregulation by transforming growth factor beta required for activation of the p15 (link4b) G(1) arrest pathway. Mol Cell Biol. 1999. 19:5913–5922.
29. Zetterberg A, Larsson O, Wiman KG. What is the restriction point? Curr Opin Cell Biol. 1995. 7:835–842.

30. Hashimoto O, Ueno T, Kimura R, Ohtsubo M, Nakamura T, Koga H, Torimura T, Uchida S, Yamashita K, Sata M. Inhibition of proteasome-dependent degradation of Wee1 in G2-arrested Hep3B cells by TGF beta 1. Mol Carcinog. 2003. 36:171–182.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download