Abstract
Promoter hypermethylation of the p16INK4a gene was investigated in 81 sets of samples of tumor tissue and adjacent normal tissue from Korean patients with primary lung cancer, using the modified real-time polymerase chain reaction (PCR)/ SYBR Green detection method. The results showed hypermethylation of p16INK4a in 27.2% of tumor tissues, and in 11.1% of adjacent normal tissue. No significant association was found between the overall aberrant methylation in tumor and corresponding normal specimens (r=0.137, p=0.219). In 22 cases with p16INK4a hypermethylation in tumor tissues, only 4 (18.1%) cases were found to have a hypermethylated normal tissue specimen. The findings of this study show that smoking can influence the methylation level of the promoter region of p16INK4a, and that this occurs in tumor tissues more frequently than in normal tissues. Other clinicopathological characteristics, including age, sex, tumor stage, and histologic type were not found to be correlated with p16INK4a methylation.
DNA methylation plays an essential role in the normal development and in the maintenance of genomic stability (1-3). However, alterations in methylation patterns frequently occur in tumor cells, and hypermethylation in the promoter regions of tumor suppressor genes are commonly associated with epigenetically mediated gene silencing in human cancer (4, 5).
The p16INK4a tumor suppressor gene encodes for a cyclin-dependent kinase inhibitor that is important for cell cycle progression, and the promoter hypermethylation of this gene is a frequent occurrence in human solid tumors (6).
In lung cancer, the p16INK4a gene hypermethylation has been detected in 17% (7) to 84% (8) of cases in different studies. This is most likely due to the various technical procedures used (9). Various methods of DNA methylation detection have been used, and these generally rely on a methylation-dependent modification of the original genomic DNA before any amplification step.
The aims of the present study were to evaluate the methylation status of the promoter region of p16INK4a in lung cancer tissue, and to analyze the relationships between methylation status and various clinicopathological parameters using a real-time quantitative polymerase chain reaction (PCR) protocol, which is a modified method of the previous developed PCR technique (10).
The study subjects were recruited from patients admitted to the Department of Thoracic and Cardiovascular Surgery at Dong-A University Hospital in Busan, Korea from March 2006 to January 2007. Tumor specimens were collected from a series of 81 non-small cell lung cancers (NSCLCs). The study design was approved by the Committee on Human Research of Dong-A University Hospital. The study subjects gave informed consent prior to participation in the study.
Immediately after surgical resection, tumor specimens and adjacent normal specimens were collected by a pathologist and stored at -80℃. DNA samples (10-20 mg) were obtained from tumor and non-tumorous tissue samples using Wizard genomic DNA purification kits (Promega, Madison, U.S.A.), according to the manufacturer's instructions.
Real-Time PCR (ABI PRISM 7000 Sequence Detection System, Applied Biosystems, Faster City, U.S.A.) was used to quantify genomic target sequences using SYBR Green 2X PCR Master Mix (Applied Biosystems) for detection. The p16INK4a gene methylation status was determined by real-time methylation specific PCR followed by restriction enzyme digestion.
One microgram of genomic DNA was incubated for 7 days at 37℃ with Msp I and Hpa II (New England BioLab, Beverly, MA, U.S.A.). When the external C in the sequence CCGG of p16INK4a is methylated, Msp I and Hpa II cannot cleave p16INK4a. However, unlike Hpa I, Msp II can cleave this sequence when the internal C residue is methylated.
Each PCR reaction mixture contained genomic DNA, 5 pM of primers, and SYBR green 2X PCR master mix (Applied Biosystems) in a final volume of 20 µL. The primer sequences and target sites are shown in the Fig. 1 and Table 1.
The standard amplification protocol consisted of an initial denaturation step for 10 min at 95℃, followed by 35 amplification cycles at 94℃ for 15 sec, each annealing temperature (67, 68 and 69℃) for 30 sec and 72℃ for 30 sec (Fig. 2 and Table 1). A standard curve was established with a 10-fold dilution series of DNA ranging from 1×100 to 1×103 ng. The DNA sample used for the standard curve was wi-38 at a known concentration (814 ng/µL) (Fig. 3). After PCR, each amplification reaction was checked using a dissociation curve.
Raw data were analyzed using the ABI 7000 System Software The methylation status in each sample was expressed as a threshold cycle (CT) ratio. The CT is the fractional cycle number at which the fluorescence signal reaches an arbitrary but defined threshold value within the early exponential phase of the reaction. CT values are proportional to the logarithm of the initial copy numbers of the target and are used to determine the initial copy numbers of samples. The CT ratio reflects methylation and was calculated as follows: CT ratio=(CT of target gene without tratment-CT of a target gene treated with Hpa II)/(CT of target gene without treatment-CT of a target gene treated with Msp I), The values of the CT ratio are between 0 and 1. In a totally non-methylated state, the CT ratio of the p16INK4a gene is 1; thus, a lower CT ratio reflects a higher level of methylation.
Human cell line wi-38 (KCLB No. 10075.1, epithelium, lung) was used as a positive control for methylated alleles. Water blanks were used as negative controls.
The association between the p16INK4a hypermethylation and clinicopathological parameters was analyzed using contingency tables. Statistical significance was evaluated using the χ2-test. The variables on the association analyses included: age, sex, histological subgroup, stage, smoking status, and tobacco consumption. Data on smoking habits were obtained by an interview. The difference was considered to be statistically significant if the p value was <0.05. The data were analyzed using the Statistical Package Service Solution software (SPSS version 12.0, Chicago, IL, U.S.A.).
In order to examine the methylation status of the p16INK4a promoter region, samples of tumor and normal adjacent tissues from 81 primary lung cancer patients were analyzed. Aberrant methylation of p16INK4a was detected in samples derived from squamous cell carcinoma (SCC), adenocarcinoma (ADC), large cell carcinoma (LCC), and bronchoalveolar carcinoma (BAC), but not from small cell lung cancer (SCLC).
Among these, the frequencies for hypermethylation of p16INK4a were 27.2% in tumor tissue and 11.1% in adjacent normal specimens. Hypermethylation of p16INK4a was more frequently identified in tumor tissues than in normal tissues with a statistical significance (p=0.009). No significant association was found between the overall aberrant methylation in tumors and the corresponding normal specimens (r=0.137, p=0.219). In 22 cases with p16INK4a hypermethylation in tumor tissues, only 4 (18.1%) cases were found to have a hypermethylated normal tissue specimen (Fig. 4).
The methylation status of the p16INK4a gene was compared with the clinicopathologic features of each NSCLC tumor (Table 2). SCC and ADC, which are major histological types of NSCLC, accounted for 91.4% (74/81) of the 81 cases examined in this work. In the SCC cases, hypermethylation was more frequently detected than in ADC cases both in tumor and in normal adjacent tissues, but the difference was not significant. Importantly, the incidence of p16INK4a methylation in tumor specimens was significantly higher in heavy smokers (>40 pack-years; p=0.045) and was more likely to be observed in those who had smoked (18/61, 29.5%) than in those who had never smoked (4 of 20, 20%), albeit without a statistical significance. Other clinical characteristics, including age, sex, and tumor stage were not correlated with p16INK4a methylation.
Several studies have described the importance of DNA methylation in human cancers and have tried to identify genes that are important for the carcinogenesis process. In particular, methylation of the p16INK4a tumor suppressor gene has been studied in various malignancies including lung cancer (11-15).
Using a SYBR Green methylation-specific PCR method, p16INK4a methylation was detected in 27.2% of lung tumor tissues. Harden et al. (7) detected methylation of the p16INK4a gene in 17% of lung cancer patients, and Liu et al. (8) observed p16INK4a gene methylation in 84% of lung cancer patients. This difference may be attributed to the different methodologies used for measuring the methylation status. Moreover, racial differences might also explain the low rate of p16INK4a hypermethylation observed in lung cancer in Koreans.
The first generation of methylation detection assays involved the digestion of genomic DNA with a methylation-sensitive restriction enzyme followed by Southern blot analysis (16). These methods are relatively straightforward, but are associated with problems such as the limited availability of informative restriction sites, false positive results (due to incomplete digestion), and a requirement for large amounts of high-molecular-weight DNA. These issues have restricted the use of these methods (17). The Second-generation technique resulted from the finding that the treatment of genomic DNA with sodium bisulfite followed by an alkaline treatment converts unmethylated cytosine to uracil, while leaving methylated cytosine residues intact (18). Moreover, methylation detection assays require gel electrophoresis and many of them employ restriction enzyme digestion, radiolabeled dNTPs or hybridization probes. These labor-intensive steps have limited the usefulness of these methods.
The strongest feature of the real-time PCR/SYBR Green detection method is the continuous optical monitoring of the progress of a fluorescent PCR reaction. Fluorescence intensity increases proportionally to the amount of PCR products. Therefore, the modified method allows conclusions to be drawn concerning the methylation status relative to a control reaction.
The results of this study demonstrate that the methylation of the p16INK4a gene is associated with tobacco smoking. The association between promoter methylation and smoking has been reported in other studies for various tumor suppressor genes in NSCLC, including p16INK4a/CDKN2A and RASSF1A (19-21). Tobacco smoke contains many carcinogens, including polyaromatic hydrocarbons, chromium, cadmium, plutonium, and nickel (22). In addition, tobacco smoke is a mucosal irritant and thus induces inflammation, which results in the generation of oxygen-free radicals. Furthermore, smoking increases the activity of DNA methyltransferase (23), which drives the de novo hypermethylation of susceptible loci (24).
Clinicopathological characteristics, including age, sex, tumor stage, and histologic type were not found to be correlated with p16INK4a methylation. A previous investigation (25) demonstrated that p16INK4a hypermethylation is a common and early event during lung carcinogenesis in general. However, the small number of subjects involved in the present study was not enough to draw a definite conclusion regarding the interaction between the gene and other related factors the testing of gene and related factor interactions, which warrants further investigation in a larger study.
Notably, in 22 cases in which p16INK4a hypermethylation was observed in tumor tissues, only 4 (18.1%) cases were found to have a hypermethylation status in normal tissues. These results suggest that tumor tissues show more p16INK4a hypermethylation than corresponding normal adjacent tissues.
In conclusion, this study shows that smoking can influence the methylation level of the promoter region of p16INK4a, and that this occurs in tumor tissues more frequently than in normal tissues. Other clinicopathological characteristics, including age, sex, tumor stage, and histologic type, were not found to be correlated with p16INK4a methylation.
Figures and Tables
Fig. 1
Methylation analysis of the p16INK4a promoter sequencing (Genebank accession number X94154). The positions of the CCGG sites are underlined. Each primer is shown in bold characters.

Fig. 2
Results of p16INK4a gene promoter methylation of normal (A) and tumor (B) lung tissues by real-time PCR. 2, 3, 5: Normal lung tissues (A); 2*, 3*, 5*: tumor lung tissues (B); 1, 4, 6: positive control (wi-38); 7: negative control (water); a: no-cut DNA amplification; b: Hpa II-cut DNA amplification; c: Msp I-cut DNA amplification. Delta Rn: the magnitude of the fluorescence signal generated during the PCR at each time point.

Fig. 3
Result of serial dilutions to determine the detection limits of the real-time PCR protocol showing the initial DNA amounts used in the amplification.

Fig. 4
Methylation status of the p16INK4a gene prompter in normal and tumor tissues. The black circle denotes methylation positivity, and the open circle indicates that the sample was negative for methylation. N, normal tissue; T, tumor tissue.
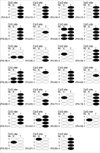
Table 1
Primer sequences and annealing temperature for PCR reactions for promoter regions of the p16INK4a gene

ACKNOWLEDGMENT
This work was supported by the Korean Science and Engineering Foundation of the MRCCMT at Dong-A University.
References
1. Monk M. Epigenetic programming of differential gene expression in development and evolution. Dev Genet. 1995. 17:188–197.

2. Turker MS, Bestor TH. Formation of methylation patterns in the mammalian genome. Mutat Res. 1997. 386:119–130.

3. Paulsen M, Ferguson-Smith AC. DNA methylation in genomic imprinting, development and disease. J Pathol. 2001. 195:97–110.

4. Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998. 72:141–196.
5. Gray SG, Eriksson T, Ekstrom TJ. Methylation, gene expression and the chromatin connection in cancer (review). Int J Mol Med. 1999. 4:333–350.

6. Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001. 61:3225–3229.
7. Harden SV, Tokumaru Y, Westra WH, Goodman S, Ahrendt SA, Yang SC, Sidransky D. Gene promoter hypermethylation in tumors and lymph nodes of stage I lung cancer patients. Clin Cancer Res. 2003. 9:1370–1375.
8. Liu Y, An Q, Li L, Zhang D, Huang J, Feng X, Cheng S, Gao Y. Hypermethylation of p16INK4a in Chinese lung cancer patients: biological and clinical implications. Carcinogenesis. 2003. 24:1897–1901.

9. Ulivi P, Zoli W, Calistri D, Fabbri F, Tesei A, Rosetti M, Mengozzi M, Amadori D. p16INK4A and CDH13 hypermethylation in tumor and serum of non-small cell lung cancer patients. J Cell Physiol. 2006. 206:611–615.
10. Chu DC, Chuang CK, Fu JB, Huang HS, Tseng CP, Sun CF. The use of real-time quantitative polymerase chain reaction to detect hypermethylation of the CpG islands in the promoter region flanking the GSTP1 gene to diagnose prostate carcinoma. J Urol. 2002. 167:1854–1858.

11. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1997. 37:646–650.
12. Nakayama H, Hibi K, Takase T, Yamazaki T, Kasai Y, Ito K, Akiyama S, Nakao A. Molecular detection of p16 promoter methylation in the serum of recurrent colorectal cancer patients. Int J Cancer. 2003. 105:491–493.

13. Reed AL, Califano J, Cairns P, Westra WH, Jones RM, Koch W, Ahrendt S, Eby Y, Sewell D, Nawroz H, Bartek J, Sidransky D. High frequency of p16 (CDKN2/MTS-1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res. 1994. 56:3630–3633.
14. Stroun M, Anker P, Maurice P, Lyautey J, Lederrey C, Beljanski M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989. 46:318–322.

15. Wong TS, Man MW, Lam AK, Wei WI, Kwong YL, Yuen AP. The study of p16 and p15 gene methylation in head and neck squamous cell carcinoma and their quantitative evaluation in plasma by real-time PCR. Eur J Cancer. 2003. 39:1881–1887.

16. Singer-Sam J, LeBon JM, Tanguay RL, Riggs AD. A quantitative HpaII-PCR assay to measure methylation of DNA from a small number of cells. Nucleic Acids Res. 1990. 18:687.
17. Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000. 28:E32.

18. Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992. 89:1827–1831.

19. Kim DH, Nelson HH, Wiencke JK, Zheng S, Christiani DC, Wain JC, Mark EJ, Kelsey KT. p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res. 2001. 61:3419–3424.
20. Jarmalaite S, Kannio A, Anttila S, Lazutka JR, Husgafvel-Pursiainen K. Aberrant p16 promoter methylation in smokers and former smokers with nonsmall cell lung cancer. Int J Cancer. 2003. 106:913–918.
21. Kim DH, Kim JS, Ji YI, Shim YM, Kim H, Han J, Park J. Hypermethylation of RASSF1A promoter is associated with the age at starting smoking and a poor prognosis in primary non-small cell lung cancer. Cancer Res. 2003. 63:3743–3746.
22. Rom WN, Hay JG, Lee TC, Jiang Y, Tchou-Wong KM. Molecular and genetic aspects of lung cancer. Am J Respir Crit Care Med. 2000. 161:1355–1367.

23. Belinsky SA, Nikula KJ, Baylin SB, Issa JP. Increased cytosine DNA-methyltransferase activity is target-cell-specific and an early event in lung cancer. Proc Natl Acad Sci USA. 1996. 93:4045–4050.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download