Abstract
Methylation of p16 is an important mechanism in cervical carcinogenesis. However, the relationship between cervical squamous cell carcinoma (SCC) and Epstein-Barr virus (EBV) remains controversial. Here, we explored whether EBV infection and/or p16 gene inactivation would play any role in cervical carcinogenesis. Eighty-two specimens included 41 invasive SCCs, 30 cervical intraepithelial neoplasm (CIN; CIN 1, 11 cases, CIN II, 3 cases, CIN III 16 cases) and 11 nonneoplastic cervices. EBV was detected by polymerase chain reaction (PCR) for EBNA-1 and in situ hybridization for EBER-1. The p16 methylation-status and the expression of p16 protein were studied by methylation-specific PCR and immunohistochemistry, respectively. The materials were divided into four groups: 1) nonneoplastic cervices, 2) CIN I, 3) CIN II-III and 4) invasive SCCs. p16 methylation and p16 immunoexpressions increased in CIN and invasive SCCs than nonneoplastic tissue. p16-methylation and p16-immunoreactivities were higher in the EBV-positive group (p=0.009, p<0.001) than in the EBV-negative group. EBV was detected more frequently in CIN and SCCs than nonneoplastic cervices. In conclusion, a correlation between p16 methylation, p16 immunoreactivity and the detection of EBV strongly suggested that the cooperation of EBV and p16 gene may play a synergic effect on cell cycle deregulation.
The p16 gene is one of the cell cycle regulating genes and encodes a nuclear protein, p16 which inhibits the D-type cyclin/cyclin-dependent kinase complexes that phosphorylate the retinoblastoma gene product (pRb), thus blocking G1-S cycle progression (1, 2). The inactivation of p16 tumor suppressor gene promotes cell proliferation, and is found in many different types of carcinomas such as gastric carcinoma, bladder tumor, glioma, breast cancer and head and neck tumors (3). There is compelling evidence that the inactivation of p16 is an important genetic event in immortalization of keratinocytes (4). In previous studies of the p16 in cervical carcinomas, methylation specific polymerase chain reaction (PCR) has shown a high level of methylation (1, 5), concordant with reports that the p16 gene is frequently inactivated through methylation rather than mutation or deletion. DNA methylation is a frequent epigenetic event in many human cancers (6, 7), and the factors inducing methylation remain unclear, but structural abnormalities of local DNA, and exposure to heavy metals are only known causes (8). A growing number of cancer-related genes are being recognized for the presence of dense methylation of cytosine in normally unmethylated CpG-rich sequences, called CpG islands within the 5' gene promoter regions.
Besides p16 gene inactivation, viral infection also participates in the dysregulation of cell cycle. A clear relationship between human papillomavirus (HPV) and cervical squamous cell carcinomas (SCCs) is well established; HPV-infected invasive SCCs tend to express more immunoreactivity for p16 protein or p16 methylation status than do HPV-negative cervical carcinomas, which is now regarded as surrogate biomarkers of HPV infection along with Ki-67 labeling index (5, 9, 10). Epstein-Barr virus (EBV) was suggested as another oncogenic virus in cervical carcinogenesis, based on findings of clonal nature of EBV in cervical carcinoma cells and the presence of EBV in precancerous lesions of the cervix (11). Since then, much debate has been evoked because of divergent results including a relatively high prevalence rate of EBV-positive nonneoplastic cervical tissue, which shed doubt on the possibility. However, heterogeneous reports on lymphoepithelioma-like carcinoma of the uterine cervix in Asian women showing a higher EBV infection (12), and the co-work of HPV and EBV cannot exclude EBV's direct, or indirect impact on cervical carcinogenesis (7, 11, 13). Considering that the tumors like gastric or nasopharyngeal carcinomas, which are known to be closely related with EBV's oncogenic effects, were identified in a high frequency of p16 methylation (14, 15), a close relationship exists between EBV and p16 gene. In Korea, one study about HPB and p16 methylation has been retrieved (16), and few studies about EBV detection or p16 alterations in cervical carcinomas have been retrieved (17, 18). There is, however, virtually no published information on the methylation changes during the multistage pathogenesis of EBV-related cervical lesions.
In this study, we explored EBV infection of the uterine cervix with reference to p16 gene that is frequently methylated in cervical carcinomas. Moreover, we investigated as to whether the two factors play an independent or synergic role during cervical carcinogenesis.
Eighty-two, formalin-fixed, paraffin-embedded cervical lesions were used for immunohistochemical, polymerase chain reaction (PCR) and in situ hybridization (ISH) studies. Materials were obtained from the pathology files of Anam Hospital, Korea University College of Medicine between 1995 and 2001. Samples included 41 cases of SCCs, 30 cases of cervical intraepithelial neoplasms (CIN 1; 11 cases, CIN II; 3 cases, CIN III; 16 cases), and 11 cases of non-neoplastic cervices.
DNA samples were extracted from several serial six µm-thick paraffin sections as previously described (19). Briefly, tissue samples were treated with lysis buffer containing 100 µg/mL proteinase K (Merck, Darmstadt, Germany) at 48℃ for 48 hr. DNA extraction was then performed by phenol/chloroform treatment and precipitation with ethanol. The quality of the DNA extracted was confirmed with a beta-globin gene-specific primer pair (BGLO1 and BGLO2), which amplifies 110 base pairs.
DNA methylation patterns in the CpG islands of the p16 were determined by MSPCR assays by Herman et al. (20). Briefly, 200 to 300 ng DNA was first treated with 3 mol/L sodium bisulfite to convert nonmethylated cytosine to uracil residues. As a diagnostic step, we used heminested PCR to increase the sensitivity of detection with primers p16M/p16 mol/L2r for the first and p16Mf/p16Mr for the second step under conditions as described. Base sequences of used primers are as follows;
p16-methylated (p16-M):
p16-unmethylated (p16-U):
In all cases, the amplification of nonmethyl-specific alleles served as controls for the efficiency of cytosine conversion and DNA quality. Controls without DNA and positive controls for U and M reactions were performed for each set of PCRs. The PCR product was visualized in agarose gels stained with ethidium bromide under ultraviolet illumination. DNA methylation was determined by the presence of a150-bp and 151-bp fragments in those samples amplified with the p16-M and p164-U primers, respectively. Positive PCR products in p16-M and/or p16-U are interpreted as methylated, and PCR products in only p16-U are interpreted as unmethylated status.
Immunohistochemical staining of p16 was performed with p16 monoclonal antibody (SC 468; Santa Cruz Biochemicals, CA, U.S.A.) diluted at 1:50. Immunodetection was performed with biotinylated antimouse immunoglobulins, followed by peroxidase-labeled streptavidin, LSAB-2 (DAKO, Glostrup, Denmark) with diaminobenzidine chromogen as the substrate. Each section was counterstained with hematoxylin. Incubation omitting the specific antibody, as well as with unrelated antibodies, was used as a control of the technique. A positive control was included with each slide. The results were interpreted as positive according to the established criteria by Geradts et al. (21).
To examine the presence of EBV DNA in the samples, 0.1 µg DNA from a cervical sample was subjected to PCR analysis using EBNA-1 primers which target a 138 base pair segment of the Bam H1W internal repeat (IR 1) region described by Coates et al. (22). Base sequences of primers were as follows;
Thirty-five PCR cycles at 95℃ for 7 min, at 94℃ for 1
min, at 58℃ for 40 sec, and at 72℃ for 40 sec were carried
out. And it was carried out at 72℃ for 7 min Polymerase
chain reaction was carried out in duplicate or triplicate. The amplified products were run on 2% Nuieve/agarose gel and were transferred onto a Hybond N+ membrane (Amersham Biosciences, Uppsala, Sweden). They were stained with ethidium bromide and examined under ultraviolet illumination. As a positive control of EBNA-1, B95-8 cell line was used.
EBV ISH was performed using oligonucleotide probes against EBV-encoded RNA-1 (EBER-1; Novocastra, Newcastle, U.K.), following previously reported procedures (23). EBER-1 positivity was shown by latently EBV-infected cells. Briefly, EBER-1 ISH was performed on each case using digoxigenin-labeled riboprobes produced from plasmid templates. A step-by-step description of one of the testing protocols, which uses EBER-1 was executed on each case and hybridized overnight. RNase was used to facilitate the washing away of unbound probe and a detection system based on the application of antibody to digoxigenin was linked to alkaline phosphatase. It was counterstained with hematoxylin. Known cases of EBV-harboring tonsils were used as controls. Strong signals of the nuclei were interpreted as positive results.
We divided the cases into four groups as follows: non-neoplastic normal cervical tissue, low grade dysplastic lesions (cervical intraepithelial neoplasm, CIN I), high grade lesions (CIN II and III) and invasive SCCs. Subsequently, we compared EBV infection with p16 methylation and p16 protein expression. Statistical analysis was performed by means of SPSS software version 10. The associations between the discrete variables were assessed using McNemar's exact test and chi-square test. The same tests were used to evaluate the influence of the tumor grade of CIN. Differences were considered statistically significant for p<0.05.
All the results of p16 methylation, p16 protein expression, EBNA-1 PCR and EBER-1 ISH of the eighty-two cases were shown in Table 1.
Non-neoplastic cervices showed unmethylation in all the cases, but 40% (12/30) of cervical intraepithelial neoplasms and 61% of invasive squamous cell carcinomas (25/41) showed p16 methylation (p=0.003, Fig. 1, Table 1). There existed statistical differences among CIN I, II and III (p=0.03, data not shown), while the advanced stage of squamous cell carcinomas (stage IIb) revealed a higher p16 methylation rate than the early stage (I, IIa) of squamous cell carcinomas, however, the results were not statistically significant (p=0.107, Table 2).
Non-neoplastic cervices were immunonegative for p16 protein except for basal cells that are known to normally express p16 protein, but 53% of cervical intraepithelial neoplasm (16/30 cases) and 68.3% of invasive squamous cell carcinomas (28/41) expressed p16 protein. These results were significantly different among the four groups (p=0.001, Table 1, Fig. 2). However, no differences existed among the three grades of cervical intraepithelial neoplasms (p=0.600, data not shown). In comparison of invasive carcinomas according to the stage, there was a higher expression rate in more advanced stages, but no statistically significant differences existed (p=0.192, Table 2).
EBV was detected by EBNA-1 PCR in 9.1% of non-neoplastic cervical tissue (1/11), 36.7% of cervical intraepithelial neoplasm (11/30) and 36.6% of invasive squamous cell carcinomas (15/41) (Fig. 3). There was no statistically significant difference in the detection rate of EBV among the four groups (p=0.331, Table 1), whereas a significant difference existed between the two groups (non-neoplastic cervices vs. precancerous CIN and cervical cancers, p=0.040, data not shown). There were no statistical differences among the grades of CIN (p=0.747, data not shown).
By McNemar's exact test, p16 MSPCR and the immunohistochemical results were proven to be concordant, while the detection rate of EBV was different between in situ hybridization and PCR.
The cervical lesions positive for EBV by PCR showed a higher rate of p16 methylation and/or p16 immunopositivity, which was statistically significant (Table 3).
In this study, 61% of squamous cell carcinomas of the uterine cervix showed p16 methylation, which was higher than 31% in the report of Wong and his colleagues (2). Cervical carcinomas are known to show a high proportion of p16 methylation, while other gynecological malignancies have variable frequencies (24). Sano et al. (4) reported that invasive squamous cell carcinoma and CIN lesions (low and high grades) exhibited higher p16 immunoreactivity than non-neoplastic cervices. Based on the results that dysplastic lesions progressed toward carcinomas with increasing p16 methylation, methylation has been thought to play a critical role in oncogenesis as in lymphoma or carcinomas of other organs (25). However, the frequency of p16 methylation between early and advanced stages showed no statistically significant difference, which is quite different from those of the tumors in the head and neck, breast or bladder (2, 26, 27). This result may indicate that early cervical carcinogenesis requires p16 methylation, but additional molecular genetic events are necessary for its progression.
Variable data on p16 immunoreactivities and cervical carcinomas have been described in the literature; most of them reported loss of p16 protein (28), while some reported the reexpression of p16 protein (29). In our study, dysplastic cervical epithelium and squamous cell carcinomas showed a higher expression of p16 protein in the cervical epithelium, which contradicts most of the previous studies, but fits well with that of Klaes et al. (29). The statistically concordance despite discrepancy in individual comparison between p16 immunohistochemistry and p16 MSPCR in this study indicates that the former may employ an inexpensive screening method to detect squamous cells showing early dysplastic changes. Interpretation of immunoreactivity for p16 protein has some limitations because p16 expression is variable in normal, dysplastic epithelium and neoplastic tissue; basal layer of nonneoplastic exocervix expresses weak and focal nuclear stainability (30). Theoretically, p16 protein should be expressed only in the nuclei but cytoplasmic staining pattern was occasionally observed, and its significance remains unclear. Cytoplasmic immunoreactivity may be induced by a nonspecific uptake or may be caused by functional heterogeneity (21). Some of the cultured cells pertaining to p16 gene show nuclear negativity but with variable cytoplasmic reactivities, or cytoplasmic p16 detected by immunoblotting may indicate that this cytoplasmic localization of p16 is "dormant" like that of the tumor suppressor gene, which has functions different from the nuclear one (31). A discrepancy in the results between the two detection methods for p16 inactivation was noted in our series. MSPCR has been known to be more sensitive and specific than Southern hybridization (20). In contrast, immunohistochemistry using p16 monoclonal antibody is simple and useful but has several technical limitations. For example, p16 protein is prone to break during the processing, and heterogeneous immunoreactivity in nonneoplastic tissue could cause confusion during interpretation. Progressive increase of p16 immunoreactivity and methylation were observed at carcinomatous progression. Timmermann et al. (32) demonstrated that epithelial cells gained increased activity of beta-galactosidase that is associated with senescence and produced p16 protein with no normal suppressor function. The reexpression of this incomplete protein detected by immnohistochemical method may be explained (33).
In this study, 60.5% of EBV-positive cervical tissues were p16-methylated, whereas 68.2% of EBV-negative cervical tissues were p16-unmethylated. These results were similar to those of nasopharyngeal undifferentiated carcinoma or EBV-positive gastric carcinomas, where there was likewise a trend toward an association between EBV infection and loss of p16 expression. One possible explanation for the association of p16 loss and EBV infection is that EBV-infected tumors could have a higher growth rate and therefore a greater opportunity to accumulate more mutations, including those of p16. In this study, we confirmed EBV infection by PCR and ISH. There was disconcordance between the results through EBV PCR and ISH. In general, the amplification of DNA through PCR was possible even in one viral particle, thus, theoretically it is a more sensitive method than ISH (9). In practice, a sensitive and specific practical method for detecting EBV is ISH (34). Cases of EBV-negativity by PCR but EBV-positive reaction by ISH in this study, seemed to be caused by insufficient amplification process. In this study, 39% of SCCs were EBV-positive, which was not a high frequency compared to non-neoplastic tissues or CIN. However, EBV-positive carcinomas showing a high rate of p16 methylation suggested that EBV alone does not play a key role in uterine cervical squamous epithelium, rather it may be involved in the earliest step of oncogenesis, which is somewhat dissimilar to that of nasopharyngeal carcinoma or Burkitt's lymphoma. Recent development of molecular genetics for carcinogenesis and virus, methylation of CpG islands possessing p16 is induced by the integration of viral DNA into host cells (34, 35). It was suggested that methylation of foreign DNA integrated with host DNA or adjacent host DNA, especially cell cycle regulatory site is the first step of carcinogenesis (36), which is similar to those of SV40 in relation to malignant mesothelioma or EBV in relation to EBV-positive gastric carcinomas (15, 37). Methylation of EBV genome inhibits suppression of immunodominant viral antigen by EBV-infected tumor cells and escapes immunosurveillance (15). Pharmacological blocking by methylation inhibitor causes tumor suppressor gene to be protected from aberrant methylation, might finally gain its own function, which the tumor cells expose immunosurveillance system. Actually, clinical application is now being attempted (38). On this mechanism, the relationship between EBV infection and p16 methylation plays a role in the early change of cervical carcinogenesis, and may provide promise for future treatments.
In conclusion, we did not find an association between p16 methylation status and stage. These observations well corresponded to the fact that p16 inactivation is an early epigenetic event in cervical cancer. A more intense reactivity of p16 protein was associated with p16 methylated status in this study, which might imply that p16 immunoreactivity represents the reactivation of senescent p16 protein in cancers, keeping with a previous report of cervical carcinoma, but differs from the correlation with the advanced stage of cervical SCC in the earliest study of Wong et al. (2). Although our findings need to be extended to a larger series, the pattern of p16 methylation in cervical lesions with or without EBV-infection may help identify subgroups at increased risk for accelerated progression.
Figures and Tables
Fig. 1
Methylation status of the p16 gene detected by methylation-specific PCR. Lane 1-3, CIN; lane 4-6, cervical carcinoma; lane 7, normal cervical tissue; M, Molecular weight marker; lanes m, reactions using p16-M primers specific for the methylated CpG sites. lanes u, reactions using p16-U primers specific for the unmethylated CpG sites.
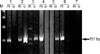
Fig. 2
Immunohistochemistry for p16. Focal nuclear staining in the basal cells of the non-neoplastic cervical tissue (A, p16 immunostain, ×200), CIN I (B, p16 immunostain, ×200), CIN III (C, p16 immunostain, ×200) and intense nuclear staining in invasive squamous cell carcinoma (D, p16 immunostain, ×400).

Fig. 3
Analysis of EBV using EBNA-1 primer. Lane 1-2, Normal cervical tissue; lane 3-6, Squamous cell carcinoma; lane 7-9, CIN; lane N, Negative control; lane C, Positive control; M, molecular weight marker.
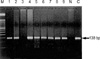
Fig. 4
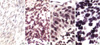
(A) EBER-1 signals detected by in situ hybridization (×200). No signals are detected in squamous cell carcinoma. (B) In CIN I, some of the dysplastic cells show nuclear staining (×200). (C) In CIN III, dysplastic cells show nuclear staining (×400). (D) In squamous cell carcinoma, tumor cells and some intervening lymphocytes show nuclear staining for EBER-1 (×400).
References
1. Nuovo GJ, Plaia TW, Belinsky SA, Baylin SB, Herman JG. In situ detection of the hypermethylation-induced inactivation of the p16 gene as an early event in oncogenesis. Proc Natl Acad Sci USA. 1999. 96:12754–12759.

2. Wong YF, Chung TK, Cheung TH, Nobori T, Yu AL, Yu J, Batova A, Lai KW, Chang AM. Methylation of p16INK4A in primary gynecologic malignancy. Cancer Lett. 1999. 136:231–235.
3. Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995. 55:4525–4530.
4. Sano T, Oyama T, Kashiwabara K, Fukuda T, Nakajima T. Expression status of p16 protein is associated with human papillomavirus oncogenic potential in cervical and genital lesions. Am J Pathol. 1998. 153:1741–1748.

5. Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001. 10:687–692.

6. Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5' CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995. 1:686–692.

7. Keating JT, Ince T, Crum CP. Surrogate biomarkers of HPV infection in cervical neoplasia screening and diagnosis. Adv Anat Pathol. 2001. 8:83–92.

8. Keating JT, Cviko A, Riethdorf S, Riethdorf L, Quade BJ, Sun D, Duensing S, Sheets EE, Munger K, Crum CP. Ki-67, cyclin E, and p16INK4 are complimentary surrogate biomarkers for human papilloma virus-related cervical neoplasia. Am J Surg Pathol. 2001. 25:884–891.
9. Landers RJ, O'Leary JJ, Crowley M, Healy I, Annis P, Burke L, O'Brien D, Hogan J, Kealy WF, Lewis FA. Epstein-Barr virus in normal, pre-malignant and malignant lesions of the uterine cervix. J Clin Pathol. 1993. 46:931–935.

10. Taylor Y, Melvin WT, Sewell HF, Flannelly G, Walker F. Prevalence of Epstein-Barr virus in the cervix. J Clin Pathol. 1994. 47:92–93.

11. Sasagawa T, Shimakage M, Nakamura M, Sakaike J, Ishikawa H, Inoue M. Epstein-Barr virus (EBV) genes expression on cervical intraepithelial neoplasia and invasive cervical cancer: a comparative study with human papillomavirus (HPV) infection. Hum Pathol. 2000. 31:318–326.
12. Noel J, Lespagnard L, Fayt I, Verhest A, Dargent J. Evidence of human papilloma virus infection but lack of Epstein-Barr virus in lymphoepithelioma-like carcinoma of uterine cervix: report of two cases and review of the literature. Hum Pathol. 2001. 32:135–138.

13. Shibosawa E, Tsutsumi K, Koizuka I, Hoshikawa M, Takakuwa T. Absence of nuclear p16 from Epstein-Barr virus-associated undifferentiated nasopharyngeal carcinomas. Laryngoscope. 2000. 110:93–97.

14. Schneider BG, Gulley ML, Eagan P, Bravo JC, Mera R, Geradts J. Loss of p16/CDKN2A tumor suppressor protein in gastric adenocarcinoma is associated with Epstein-Barr virus and anatomic location in the body of the stomach. Human Pathol. 2000. 31:45–50.

15. Kang GH, Lee S, Kim WH, Lee HW, Kim JC, Rhyu MG, Ro JY. Epstein-barr virus-positive gastric carcinoma demonstrates frequent aberrant methylation of multiple genes and constitutes CpG island methylator phenotype-positive gastric carcinoma. Am J Pathol. 2002. 160:787–794.

16. Cho NH, Kim YT, Kim JW. Alteration of cell cycle in cervical tumor associated with human Papillomavirus: Cyclin-dependent kinase inhibitors. Yonsei Med J. 2002. 43:722–728.

17. Kim IS, Kang JS, Choi AN, Kim YS. The prevalence of Epstein-Barr virus in uterine cervical cancer: Detection by PCR and in situ PCR methods. Korean J Obstet Gynecol. 2000. 43:184–191.
18. Jeong HJ, Lee ES, Lin ZH, Park SH, Kim IS, Kang JS. Detection of Epstein-Barr virus in the inflammatory and neoplastic uterine cervical lesions. Korean J Cytopathol. 2001. 12:73–80.
19. Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987. 19:11–15.
20. Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad USA. 1996. 93:9821–9826.

21. Geradts J, Kratzke RA, Niehans GA, Lincoln CE. Immunohistochemical detection of the cyclin-dependent kinase inhibitor 2/multiple tumor suppressor gene 1 (CDKN2/MTS1) product p16INK4A in archival human solid tumors: correlation with retinoblastoma protein expression. Cancer Res. 1995. 55:6006–6011.
22. Coates PJ, d'Ardenne AJ, Khan G, Kangro HO, Slavin G. Simplified procedures for applying the polymerase chain reaction to routinely fixed paraffin wax sections. J Clin Pathol. 1991. 44:115–118.

23. Chang KL, Chen YY, Shibata D, Weiss LM. Description of an in situ hybridization methodology for detection of Epstein-Barr virus RNA in paraffin-embedded tissues, with a survey of normal and neoplastic tissues. Diagn Mol Pathol. 1992. 1:246–255.

24. Virmani AK, Muller C, Rathi A, Zoechbauer-Mueller S, Mathis M, Gazdar AF. Aberrant methylation during cervical carcinogenesis. Clin Cancer Res. 2001. 7:584–589.
25. Wong YF, Chung TK, Cheung TH, Nobori T, Yim SF, Lai KW, Phil M, Yu AL, Diccianni MB, Li TZ, Chang AM. p16INK4 and p15INK4B alterations in primary gynecologic malignancy. Gynecol Oncol. 1997. 65:319–324.
26. Salem C, Liang G, Tsai YC, Coulter J, Knowles MA, Feng AC, Groshen S, Nichols PW, Jones PA. Progressive increases in de novo methylation of CpG islands in bladder cancer. Cancer Res. 2000. 60:2473–2476.
27. Yuen PW, Man M, Lam KY, Kwong YL. Clinicopathological significance of p16 gene expression in the surgical treatment of head and neck squamous cell carcinomas. J Clin Pathol. 2002. 55:58–60.
28. Villuendas R, Sanchez-Beato M, Martinez JC, Saez AI, Martinez-Delgado B, Garcia JF, Mateo MS, Sanchez-Verde L, Benitez J, Martinez P, Piris MA. Loss of p16/INK4A protein expression in non-Hodgkin's lymphomas is a frequent finding associated with tumor progression. Am J Pathol. 1998. 153:887–897.

29. Klaes R, Friedrich T, Spitkovsky D, Ridder R, Rudy W, Petry U, Dallenbach-Hellweg G, Schmidt D, von Knebel Doeberitz M. Overexpression of p16INK4A as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int J Cancer. 2001. 92:276–284.
30. Nielsen GP, Stemmer-Rachamimov AO, Shaw J, Roy JE, Koh J, Louis DN. Immunohistochemical survey of p16INK4A expression in normal human adult and infant tissues. Lab Invest. 1999. 79:1137–1143.
31. Shiozawa T, Nikaido T, Shimizu M, Zhai Y, Fujii S. Immunohistochemical analysis of the expression of cdk4 and p16INK4 in human endometrioid-type endometrial carcinoma. Cancer. 1997. 80:2250–2256.
32. Timmermann S, Hinds PW, Munger K. Re-expression of endogenous p16ink4a in oral squamous cell carcinoma lines by 5-aza-2'-deoxycytidine treatment induces a senescence-like state. Oncogene. 1998. 17:3445–3453.
33. Gump J, Koh J. An antibody to p16INK4A recognizes a modified form of galectin-3. Hybridoma. 2001. 20:167–174.
34. Takeuchi H, Kobayashi R, Hasegawa M, Hirai K. Detection of latent Epstein-Barr virus (EBV) DNA in paraffin sections of nasopharyngeal carcinomas expressing no EBV-encoded small RNAs using in situ PCR. Arch Virol. 1997. 142:1743–1756.

35. Remus R, Kammer C, Heller H, Schmitz B, Schell G, Doerfler W. Insertion of foreign DNA into an established mammalian genome can alter the methylation of cellular DNA sequences. J Virol. 1999. 73:1010–1022.

36. Klein CB, Costa M. DNA methylation, heterochromatin and epigenetic carcinogens. Mutat Res. 1997. 386:163–180.

37. Toyooka S, Pass HI, Shivapurkar N, Fukuyama Y, Maruyama R, Toyooka KO, Gilcrease M, Farinas A, Minna JD, Gazdar AF. Aberrant methylation and simian virus 40 tag sequences in malignant mesothelioma. Cancer Res. 2001. 61:5727–5730.
38. Reid GK, Besterman JM, MacLeod AR. Selective inhibition of DNA methyltransferase enzymes as a novel strategy for cancer treatment. Curr Opin Mol Ther. 2002. 4:130–137.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download