The acquisition of multidrug resistance in nosocomial pathogens such as Serratia marcescens has been reported in the absence of antibiotic selection pressure. The broad antimicrobial spectrum of aminoglycosides and their synergistic antimicrobial effect in association with other drugs (such as β-lactams and fluoroquinolones) suggest the possible use of these antibiotics as a therapy for life-threatening infections. One of the most important mechanisms of aminoglycoside resistance is posttranscriptional methylation of 16S rRNA conferred by methyltransferases. To date, 10 kinds of 16S rRNA methyltransferase genes (armA, rmtA, rmtB, rmtC, rmtD, rmtE, rmtF, rmtG, rmtH, and npmA), whose products confer high-level resistance to aminoglycosides, have been reported in various Enterobacteriaceae strains [1]. The rmtB gene was first identified in S. marcescens in Japan, but has not been reported in S. marcescens elsewhere [2]. In the present study, we described an isolate of S. marcescens harboring rmtB, together with quinolone resistance genes and various β-lactamase genes, for the first time in China.
A strain of S. marcescens, designated GN1384, was isolated from the urine of a 81-yr-old male patient who suffered from urinary tract infections in September 2009 and was admitted to the second hospital of Hefei, China. Species identification was performed with the Vitek2 system (bioMérieux, Marcy l'Étoile, France) and confirmed with API 20E (bioMérieux). The minimum inhibitory concentrations (MICs) of various antibiotics were determined by using the agar dilution method, and susceptibility data were interpreted by using the Clinical and Laboratory Standards Institute guidelines [3]. The isolate showed a multiple drug resistant pattern, including some β-lactams, all the aminoglycosides, and fluoroquinolones (Table 1).
Then, genomic and plasmid DNA of strain GN1384 was extracted. Using primers described previously, we identified the resistance genotype of this strain by detecting the following resistance determinants through PCR, based on the above resistance phenotypes: 16S rRNA methyltransferase genes (armA, rmtA, rmtB, rmtC, rmtD, rmtE, and npmA); plasmid-mediated quinolone resistance determinants (qnrA, qnrB, qnrS, qnrC, qnrD, aac (6')-Ib-cr, and qepA); chromosome mutations associated with fluoroquinolone resistance (gyrA, parC, and parE); and genes coding for β-lactamases (TEM, CTX-M, SHV, OXA-1) [4,5,6,7]. The positive amplicons were subsequently sequenced, and the results indicated the presence of rmtB, aac(6')-Ib-cr, blaCTX-M-14, blaTEM-1, and blaOXA-1. In addition, two amino acid changes (Ser83Leu and Asp87Asn) in GyrA and five amino acid changes (Thr57Ser, Ser80Ile, Ala129Ser, Val131Leu, and Gly134Ser) in ParC were observed.
Conjugation experiments were performed with Escherichia coli J53 as the recipient, as previously described [8]. The transconjugants were selected on agar plates containing sodium azide (100 mg/L) supplemented with amikacin (128 mg/L). Conjugation experiments showed that aac(6')-Ib-cr, blaTEM-1, blaCTX-M-14, and blaOXA-1 were cotransferred with rmtB to the recipient. The MIC results revealed that it simultaneously exhibited an elevated level of resistance to β-lactams, aminoglycosides, and fluoroquinolones (Table 1).
The extremely high MICs of all aminoglycosides tested against the multidrug-resistant S. marcescens were likely due to the presence of the rmtB gene. In contrast to the rmtB plasmid isolated by Doi et al. [2], the plasmid bearing rmtB in our study was self-transferable, as observed by Bogaerts et al. [9]. In accordance with a previous study, the close linkage between 16S rRNA methyltransferase and β-lactamase as well as plasmid-mediated quinolone resistance determinants revealed the significant relationship of these genes amongst Enterobacteriaceae [10]. It was also observed that the plasmid-mediated quinolone resistance gene together with the mutations in gyrA and parC contributed to the high-level quinolone resistance in S. marcescens GN1384. Further studies are required to assess the genetic environment of these genes and the role of these new substitutions in resistance to fluoroquinolones. The continuous and widespread use of aminoglycosides, fluoroquinolones, and β-lactams in human or animal infection is the major driving force leading to these sophisticated resistance gene linkages. The dissemination of these linked genes by horizontal transfer simultaneously is very worrisome, and continuous monitoring should be reinforced.
Acknowledgments
This work was supported by a research grant from the National Natural Science Foundation of China (no. 81101288 and 8117 2737).
References
1. O'Hara JA, McGann P, Snesrud EC, Clifford RJ, Waterman PE, Lesho EP, et al. Novel 16S rRNA Methyltransferase RmtH produced by Klebsiella pneumoniae associated with war-related trauma. Antimicrob Agents Chemother. 2013; 57:2413–2416. PMID: 23478957.
2. Doi Y, Yokoyama K, Yamane K, Wachino J, Shibata N, Yagi T, et al. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob Agents Chemother. 2004; 48:491–496. PMID: 14742200.
3. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. Twenty-third informational supplement, M100-S23. Wayne, PA: CLSI;2013.
4. Berçot B, Poirel L, Nordmann P. Updated multiplex polymerase chain reaction for detection of 16S rRNA methylases: high prevalence among NDM-1 producers. Diagn Microbiol Infect Dis. 2011; 71:442–445. PMID: 22000158.

5. Barguigua A, El Otmani F, Talmi M, Bourjilat F, Haouzane F, Zerouali K, et al. Characterization of extended-spectrum ß-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates from the community in Morocco. J Med Microbiol. 2011; 60:1344–1352. PMID: 21546559.
6. Park YJ, Yu JK, Lee S, Oh EJ, Woo GJ. Prevalence and diversity of qnr alleles in AmpC-producing Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii and Serratia marcescens: a multicentre study from Korea. J Antimicrob Chemother. 2007; 60:868–871. PMID: 17660263.
7. Dutta S, Kawamura Y, Ezaki T, Nair GB, Iida K, Yoshida S. Alteration in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in Quinolone-resistant Shigella dysenteriae serotype I clinical isolates from Kolkata, India. Antimicrob Agents Chemother. 2005; 49:1660–1661. PMID: 15793166.
8. Davis MA, Baker KN, Orfe LH, Shah DH, Besser TE, Call DR. Discovery of a gene conferring multiple-aminoglycoside resistance in Escherichia coli. Antimicrob Agents Chemother. 2010; 54:2666–2669. PMID: 20368404.
9. Bogaerts P, Galimand M, Bauraing C, Deplano A, Vanhoof R, De Mendonca R, et al. Emergence of ArmA and RmtB aminoglycoside resistance 16S rRNA methylases in Belgium. J Antimicrob Chemother. 2007; 59:459–464. PMID: 17224412.

10. Li DX, Zhang SM, Hu GZ, Wang Y, Liu HB, Wu CM, et al. Tn3-associated rmtB together with qnrS1, aac(6')-Ib-cr and blaCTX-M-15 are co-located on an F49:A-:B- plasmid in an Escherichia coli ST10 strain in China. J Antimicrob Chemother. 2012; 67:236–238. PMID: 22010207.
Table 1
Characteristics of clinical isolate S. marcescens GN1384 and its transconjugant
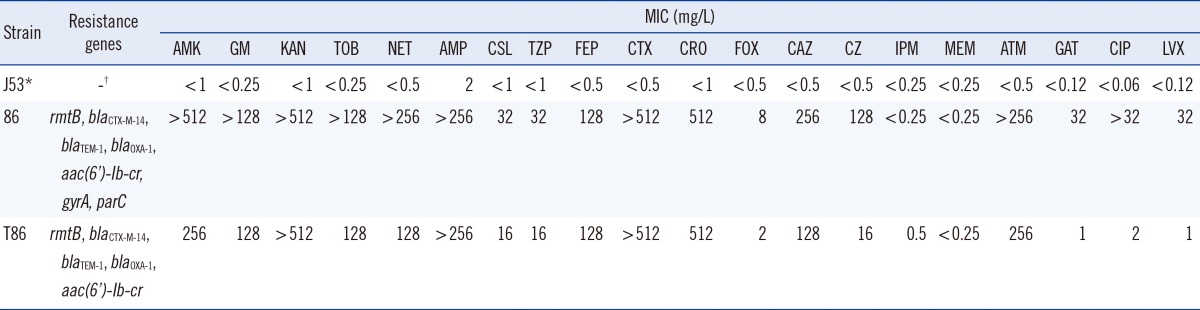
*Sodium azide-resistant Escherichia coli J53; †Indicates no genes or no information needed.
Abbreviations: AMK, amikacin; GM, gentamicin; KAN, kanamycin; TOB, tobramycin; NET, netilmicin; AMP, ampicillin; CSL, cefoperazone-sulbactam; TZP, piperacillin-tazobactam; FEP, cefepime; CTX, cefotaxime; CRO, ceftriaxone; FOX, cefoxitin; CAZ, ceftazidime; CZ, cefizoxime; IMP, imipenem; MEM, meropenem; ATM, aztreonam; GAT, gatifloxacin; CIP, ciprofloxacin; LVX, levofloxacin.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download