Abstract
Background
A peptide nucleic acid (PNA) probe-based real-time PCR (PNAqPCR™ TB/NTM detection kit; PANAGENE, Korea) assay has been recently developed for the simultaneous detection of Mycobacterium tuberculosis complex (MTBC) and nontuberculous mycobacteria (NTM) in clinical specimens. The study was aimed at evaluation of the performance of PNA probe-based real-time PCR in respiratory specimens.
Methods
To evaluate potential cross-reactivity, the extracted DNA specimens from Mycobacterium species and non-mycobacterial species were tested using PNA probe-based real-time PCR assay. A total of 531 respiratory specimens (482 sputum specimens and 49 bronchoalveolar washing fluid specimens) were collected from 230 patients in July and August, 2011. All specimens were analyzed for the detection of mycobacteria by direct smear examination, mycobacterial culture, and PNA probe-based real-time PCR assay.
Results
In cross-reactivity tests, no false-positive or false-negative results were evident. When the culture method was used as the gold standard test for comparison, PNA probe-based real-time PCR assay for detection of MTBC had a sensitivity and specificity of 96.7% (58/60) and 99.6% (469/471), respectively. Assuming the combination of culture and clinical diagnosis as the standard, the sensitivity and specificity of the new real-time PCR assay for detection of MTBC were 90.6% (58/64) and 99.6% (465/467), respectively. The new real-time PCR for the detection of NTM had a sensitivity and specificity of 69.0% (29/42) and 100% (489/489), respectively.
Tuberculosis caused by the Mycobacterium tuberculosis complex (MTBC), which affects about 8.4 million patients and causes over 1.5 million deaths annually, is a major global health problem [1]. Nontuberculous mycobacteria (NTM) appear to be distributed widely in the environment, and about one-third of NTM species have been associated with human diseases [2, 3]. Clinical diseases caused opportunistically by NTM are being encountered with an increasing frequency in AIDS and non-AIDS populations [4, 5]. Therefore, rapid and accurate identification of mycobacteria in the clinical setting is essential for the control of the spread of tuberculosis and for adequate antimicrobial therapy against mycobacterial infection [6]. Clinical microbiology laboratories have a central role in patient treatment and disease control, but conventional methods used in microbiology laboratories can pose major limitations. Culture methods for detecting mycobacteria in clinical specimens require long incubation times because of slow growth of the organisms [7]. Acid-fast bacilli (AFB) smear provides rapid results and is widely used in clinical laboratories. However, the AFB smear has low sensitivity for laboratory diagnosis because of the necessity for large numbers of organisms in a specimen for a positive result and yields poor positive predictive value for tuberculosis in clinical settings in which NTM is frequently isolated [8-10].
Since the introduction of nucleic acid amplification (NAA)-based tests with advances in genetic technologies in the recent decades, a remarkable improvement has occurred in the direct detection of mycobacteria [7]. A variety of NAA-based assays has been commercially developed for detection of MTBC and NTM from clinical specimens and are now widely used in clinical microbiology laboratories. In particular, the use of real-time PCR assay for the detection of microorganisms has been increasing, replacing conventional PCR that uses agarose gel electrophoresis for identification of PCR products. Real-time PCR assays using fluorescence resonance energy transfer (FRET) probes, molecular beacons, or TaqMan probes have been adapted for continuous detection of amplification products in a closed system. These assays have the advantage of a low contamination risk and simultaneous identification of multiple targets [11, 12].
Peptide nucleic acids (PNA) are artificially synthesized DNA analogues with an uncharged peptide backbone [13]. PNA have more favorable hybridization properties and chemical, thermal, and biological stability because of their uncharged nature and their peptide bond-linked backbone [14]. Because of these favorable characteristics, PNA has been widely applied as a diagnostic tool in molecular biology. Recently, a PNA probe-based real-time PCR assay (PNAqPCR™ TB/NTM detection kit; PANAGENE, Daejeon, Korea) was developed for the simultaneous detection of MTBC and NTM in clinical specimens. The aim of this study was to evaluate the performance of PNA probe-based real-time PCR assay in respiratory specimens.
To evaluate potential cross-reactivity, the extracted DNA specimens from 6 reference strains (M. tuberculosis [ATCC 27294], M. avium [ATCC 15769], M. intracellulare [Korean Collection for Type Culture (KCTC) 9514], M. fortuitum [KCTC 1122], Nocardia asteroides [KCTC 9956], and Rhodococcus equi [KCTC 9082]) and clinically isolated strains from respiratory specimens (Corynebacterium striatum, Klebsiella pneumoniae, Pseudomonas aeruginosa, Streptococcus pneumoniae, Staphylococcus aureus, Staphylococcus epidermidis, Moraxella species, Escherichia coli, and Acinetobacter baumannii) were tested by PNA probe-based real-time PCR assay.
A total of 531 respiratory specimens (482 sputum samples and 49 bronchoalveolar washing fluid specimens) were collected from 230 patients with suspected mycobacterial infection in July and August, 2011. All specimens were analyzed for the detection of mycobacteria by direct smear examination, mycobacterial culture, and the PNA probe-based real-time PCR assay.
All respiratory specimens were liquefied and decontaminated with N-acetyl-L-cysteine-sodium hydroxide and concentrated by centrifugation at 3,000×g for 30 min. Following concentration and resuspension of the sediments in 1.5 mL of phosphate buffer, part of the sediment from each specimen was used for AFB smear and inoculated in a BACTEC™ MGIT™ 960 system (Becton Dickinson Diagnostic Instrument Systems, Sparks, MD, USA). The remaining portion of the sediment was stored at -80℃ until the NAA assays were performed.
Smears were stained with the auramine-rhodamine fluorescent stain, and auramine-rhodamine-positive smears were confirmed by Ziehl-Neelsen staining. After a 500-µL aliquot of processed sediment was inoculated, the BACTEC™ MGIT™ 960 culture was incubated for 6 weeks at 36℃. A positive culture was confirmed by AFB staining, immunochromatographic assay kit (BIOLINE SD TB Ag MPT64 Rapid, Standard Diagnostics, Yongin-si, Korea), and PCR assay (Seeplex MTB/NTM ACE Detection, Seegene, Seoul, Korea).
The sample DNA was extracted using an InstaGene matrix (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer's instructions. Decontaminated specimens were washed with Dulbecco's phosphate-buffered saline (WelGENE, Daegu, Korea). Specimens were subjected to 5 min of centrifugation at 12,000×g. The supernatant was discarded, and the sediment was resuspended in 100 µL of InstaGene Matrix and incubated at 56℃ for 15 min. The mixtures were vortexed, incubated in a dry-heat block at 100℃ for 8 min, and centrifuged to sediment the matrix. Five microliters of each DNA sample was used as a template for amplification in real-time PCR.
The PNAqPCR™ TB/NTM detection kit includes a primer set targeting the IS6110 insertion sequence for detection of MTBC and a primer set targeting the internal transcribed spacer (ITS) sequence for detection of mycobacteria. The MTBC-specific PNA probe was labeled with Texas Red and Dabsyl (dimethyl-aminoazosulfonic acid), the mycobacteria-specific PNA probe with FAM (6-carboxyfluorescein) and Dabcyl (4,4-dimethyl-amino-azobenzene-4'-carboxylic acid), and the internal control PNA probe with HEX (4, 4, 7, 2', 4', 5', 7'-hexachloro-6-carboxyfluorescein) and Dabcyl. The primer/probe sequences cannot be revealed because of the manufacturer's copyright policy. Generation of fluorescence signals during hybridization in real-time PCR is illustrated in Fig. 1 (provided technical data from manufacturer). PNA-based real-time PCR assay was conducted in accordance with the manufacturer's instructions using a CFX96 (Bio-Rad Laboratories). Positive and negative controls were used for every reaction. For amplification, 5 µL of extracted DNA, 10 µL of the primer/probe mixture (solution A), and 10 µL of the real-time PCR master mixture (solution B) were mixed in a 96-well plate. The cycling program was 2 min at 50℃, 15 min at 95℃, and 45 cycles of 10 sec at 95℃, 30 sec at 58℃ and 15 sec at 72℃. A positive result for IS6110 and internal control was defined as a threshold cycle (CT) value≤40, and a positive result for ITS was defined as a CT value≤42. The result of an assay was regarded as invalid if the assays for IS6110, ITS, and internal control all showed simultaneously negative results. When invalid results for real-time PCR assay were obtained, the assay was repeated using a 2-fold dilution of the extracted DNA. The result of an assay was interpreted as positive for MTBC and NTM according to the manufacturer's instructions. The result of an assay was considered as positive for MTBC if the assay only showed positive results for IS6110. When the assay only showed positive result for ITS, the result was regarded as positive for NTM. When the assay showed simultaneously positive results for IS6110 and ITS, the result of the assay was interpreted as following: (i) if the CT value for ITS>the CT value for IS6110 +2, the result of the assay was considered as positive for MTBC (ii) if the CT value for ITS≤the CT value for IS6110 +2, the result was considered as simultaneously positive for MTBC and NTM.
To identify the NTM species, direct sequencing of all NTM isolates was performed. Purified DNA was amplified by using a specific primer for the Mycobacterium genus. The cycling program was as follows: 10 min at 95℃, and 30 cycles of 30 sec at 95℃, 30 sec at 65℃ and 60 sec at 72℃. The sense primer was ITS-F (5'-TGGATCCGACGAAGTCGTAACAAGG-3'), and the antisense primer was PAN-04R (5'-ATGCTCBCAABCACTATCCA-3') [15]. PCR products were purified by using a LaboPass™ PCR purification kit according to the manufacturer's instructions. Purified products were sequenced with selected amplification primers. The purified products were analyzed with an ABI 3730xl DNA analyzer (Applied Biosystems, Foster City, CA, USA).
For clinical assessment of tuberculosis, each patient's clinical records and chest radiography/computed tomography images were reviewed. Two categories of specimens were considered as true-positive specimens for tuberculosis: (i) culture-positive specimens for MTBC and (ii) samples that were culture-negative for MTBC but belonged to patients whose clinical history and radiography/computed tomography findings provided enough evidence of tuberculosis to initiate antituberculous chemotherapy.
Of the 531 respiratory specimens from 230 patients, 42 specimens were AFB smear positive and scored trace to 4+: (i) 11 specimens had trace levels, (ii) 3 specimens scored 1+, (iii) 5 specimens scored 2+, (iv) 19 specimens scored 3+, and (v) 4 specimens scored 4+. Among mycobacterial isolates in cultures from 102 clinical specimens, 60 were identified as MTBC and 42 as NTM. In the AFB smear-positive specimens, MTBC was isolated in 36 specimens that scored trace to 4+: (i) 8 specimens had trace levels, (ii) 3 specimens scored 1+, (iii) 3 specimens scored 2+, (iv) 18 specimens scored 3+, and (v) 4 specimens scored 4+. NTM was isolated in 6 smear-positive samples that scored trace to 3+: (i) 3 specimens had trace levels, (ii) 2 specimens scored 2+, and (iii) 1 specimen scored 3+. In the smear-negative specimens, MTBC was isolated in 24 specimens and NTM was isolated in 36 specimens. Of 429 smear-negative and culture-negative specimens, 4 specimens were considered to show positive results for tuberculosis by clinical diagnosis.
In the cross-reactivity test, extracted DNA from M. tuberculosis tested positive with both MTBC-specific and mycobacteria-specific PNA probes in the real-time PCR assay. Specimens extracted from M. avium, M. intracellulare, and M. fortuitum tested positive with the mycobacteria-specific PNA probe but negative with the MTBC-specific PNA probe in the real-time PCR assay. None of the specimens extracted from 11 non-mycobacterial strains tested positive with MTBC-specific or mycobacteria-specific PNA probes in real-time PCR assays.
Among the 531 clinical specimens, 88 yielded positive results in the PNA probe-based real-time PCR assay. Real-time PCR assays for 29 specimens only showed positive results for IS6110 and assays for 28 specimens only showed positive results for ITS. The assays for 31 specimens showed simultaneously positive results for IS6110 and ITS. The CT values of real-time PCR for IS6110 ranged from 22.7 to 37.7 with a median of 29.3 and the CT values for ITS ranged from 26.2 to 41.5 with a median of 35.8. The interpretation of the results of the real-time PCR assay showed positive results for MTBC in 60 specimens and positive results for NTM in 29 specimens. Among those specimens, only 1 was simultaneously positive for MTBC and NTM (Table 1).
Among the smear-positive and MTBC culture-positive specimens, all specimens showed MTBC positivity in the real-time PCR assay. For the 24 specimens that were smear-negative and MTBC culture-positive, the real-time PCR assay showed MTBC-positive results in 22 specimens. When MTBC culture-negative specimens were tested, the real-time PCR assay showed MTBC-positive results in 2 specimens, and in these samples, NTM was isolated by culture. When the culture method was used as the gold standard test for comparison, PNA probe-based real-time PCR assay for detection of MTBC had a sensitivity and specificity of 96.7% (58/60) and 99.6% (469/471), respectively. Among the 4 specimens from patients clinically diagnosed with tuberculosis that were smear-negative and MTBC culture-negative, none showed MTBC positivity in the real-time PCR assay. Assuming the combination of culture and clinical diagnosis as the standard, the sensitivity and specificity of the PNA probe-based real-time PCR assay were 90.6% (58/64) and 99.6% (465/467), respectively (Table 2). The CT values of real-time PCR for IS6110 in the MTBC-positive specimens ranged from 22.7 to 37.7 with a median of 29.1. The median CT values in smear-positive and smear-negative specimens were 27.4 and 30.7, respectively.
Among all respiratory specimens, 29 were positive for NTM by the PNA probe-based real-time PCR assay. NTM was isolated by culture in all PCR-positive specimens. However, among the 502 specimens that were NTM-negative by the real-time PCR assay, NTM was isolated by culture in only 13 specimens. The sensitivity and specificity of real-time PCR assay for detection of NTM were 69.0% (29/42) and 100% (489/489), respectively. When 6 specimens that were smear-positive and NTM culture-positive were tested, the real-time PCR assay showed NTM-positive results in 5 samples and an NTM-negative result in 1 sample with a trace smear (Table 2). The CT values obtained in real-time PCR for ITS in the NTM-positive specimens ranged from 29.7 to 41.5 with a median of 37.5. The median CT values in smear-positive and smear-negative specimens were 35.3 and 37.9, respectively. The nontuberculous mycobacterial isolates from clinical specimens and the real-time PCR results are listed in Table 3.
In this study, MTBC- and mycobacteria-specific PNA probes were used in real-time PCR for detection of tuberculosis and NTM infection. PNA probes contain random coil structures on their ends with a fluorophore and a quencher in close proximity, and fluorescence signals are inhibited by quenching. During hybridization with template DNA, PNA probes with random coil conformations straighten, resulting in increased fluorescence signals (Fig. 1). Compared with DNA probes, PNA probes have the advantages of high affinity and sequence specificity for binding to complementary nucleic acids. An important property of PNA is its uncharged nature, because the PNA backbone is composed of repeating N-(2-aminoethyl)-glycine units linked by peptide bonds instead of the negatively charged sugar phosphate backbone of natural nucleic acids [16]. The uncharged nature of PNA allows the formation of a strong PNA/DNA duplex.
The new real-time PCR assay using PNA probes for detection of MTBC revealed high sensitivity and specificity in this study. Although the new real-time PCR assay could not detect culture-negative tuberculosis, this can be attributed to the inadequate quality of specimens in the culture-negative tuberculosis samples. The performance of the new real-time PCR assay for detection of MTBC is comparable with those of other real-time PCR assays used in Korea [17]. Rapid identification of MTBC in smear-negative samples as well as in smear-positive samples is important for prevention of tuberculosis transmission, because about 17% of tuberculosis cases involve transmission from persons with negative AFB smear results [18]. Systematic reviews and meta-analyses of the performance of NAA tests for the diagnosis of tuberculosis reported that sensitivity was 96% in smear-positive samples and 66-73% in smear-negative samples [19, 20]. The sensitivity of the new real-time PCR assay in smear-negative samples is comparable with those of other NAA tests.
Unlike M. tuberculosis, no animal-to-human or human-to-human transmission of NTM has been reported. Infection of NTM is assumed to be acquired from environmental sources [4, 5]. There has been a significant rise in human disease caused by NTM during recent decades, with the increasing population of immunocompromised patients [21, 22]. Rapid identification and discrimination of NTM from MTBC is useful for the management of mycobacterial disease, because many NTM are resistant to the antibiotics used for the treatment of tuberculosis [15]. The sensitivity of the new real-time PCR for detection of NTM was significantly lower than that for detection of MTBC. We assume the reason for this low sensitivity is that the ITS sequence was used as the target for detection of mycobacteria, whereas the highly repetitive IS6110 sequence, which is present in 10 to 16 copies in most MTBC members isolated from clinical specimens, was employed for detection of MTBC [7].
The detection limit of the new real-time PCR assay was not determined in this study. Moreover, extra-pulmonary specimens were not included in our study, because a large number of specimens are required for a sufficient amount of isolated mycobacteria. Thus, further studies may be necessary to validate the new real-time PCR assay by determination of the detection limit and testing using a large number of clinical specimens in a variety of clinical settings.
In conclusion, the PNA probe-based real-time PCR assay may be useful for detection of MTBC in respiratory specimens, including smear-negative specimens, because of its high sensitivity and specificity. Further, the new real-time PCR assay may be useful for discrimination of NTM from MTBC due to its high specificity.
Figures and Tables
Fig. 1
PNA probes that contain random coil structures undergo quenching of fluorescence. Fluorescence signals are increased by the conformational change of probes during hybridization.
Abbreviation: PNA, peptide nucleic acids.
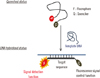
Table 1
Threshold cycle (CT) values of PNA probe-based real-time PCR assay for Mycobacterium tuberculosis complex and nontuberculous mycobacteria from respiratory specimens

Acknowledgement
This work was supported in part by the Soonchunhyang University Research Fund. We thank Young Ho Kim, Tae Young Lee and Young Il Park for excellent technical assistance.
References
1. WHO Report 2010 global tuberculosis control. World Health Organization. Updated on Sep 2011. http://whqlibdoc.who.int/publications/2010/9789241564069_eng.pdf.
2. Peter-Getzlaff S, Lüthy J, Böddinghaus B, Böttger EC, Springer B. Development and evaluation of a molecular assay for detection of nontuberculous mycobacteria by use of the cobas amplicor platform. J Clin Microbiol. 2008. 46:4023–4028.

3. Primm TP, Lucero CA, Falkinham JO 3rd. Health impacts of environmental mycobacteria. Clin Microbiol Rev. 2004. 17:98–106.

4. Richter E, Brown-Elliot BA, Wallace RJ. Versalovic J, Carroll KC, Funke G, Jorgensen JH, Landry ML, Warnock DW, editors. Mycobacterium: laboratory characteristics of slowly growing mycobacteria. Manual of clinical microbiology. 2011. 10th ed. Washington, DC: ASM Press;503–524.
5. Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007. 175:367–416.

6. Shinnick TM, Iademarco MF, Ridderhof JC. National plan for reliable tuberculosis laboratory services using a systems approach. Recommendations from CDC and the Association of Public Health Laboratories Task Force on Tuberculosis Laboratory Services. MMWR Recomm Rep. 2005. 54:1–12.
7. Forbes EA. Persing DH, Tenover FC, Tang YW, Nolte FS, Hayden RT, van Belkum A, editors. Molecular detection and characterization of Mycobacterium tuberculosis. Molecular microbiology: diagnostic principles and practice. 2011. 2nd ed. Washington, DC: ASM Press;415–436.
8. Apers L, Mutsvangwa J, Magwenzi J, Chigara N, Butterworth A, Mason P, et al. A comparison of direct microscopy, the concentration method and the Mycobacteria Growth Indicator Tube for the examination of sputum for acid-fast bacilli. Int J Tuberc Lung Dis. 2003. 7:376–381.
9. Fitzgerald DW, Sterling TR, Haas DW. Mandell GL, Bennett JE, Dolin R, editors. Mycobacterium tuberculosis. Mandell, Douglas, and Bennett's Principles and practice of infectious disease. 2010. 7th ed. Philadelphia: Churchill Livingstone Elsevier;3129–3163.

10. Centers for Disease Control and Prevention. Updated guidelines for the use of nucleic acid amplification tests in the diagnosis of tuberculosis. MMWR Morb Mortal Wkly Rep. 2009. 58:7–10.
11. Drouillon V, Delogu G, Dettori G, Lagrange PH, Benecchi M, Houriez F, et al. Multicenter evaluation of a transcription-reverse transcription concerted assay for rapid detection of Mycobacterium tuberculosis complex in clinical specimens. J Clin Microbiol. 2009. 47:3461–3465.
12. Kim K, Lee H, Lee MK, Lee SA, Shim TS, Lim SY, et al. Development and application of multiprobe real-time PCR method targeting the hsp65 gene for differentiation of Mycobacterium species from isolates and sputum specimens. J Clin Microbiol. 2010. 48:3073–3080.
13. Porcheddu A, Giacomelli G. Peptide nucleic acids (PNAs), a chemical overview. Curr Med Chem. 2005. 12:2561–2599.

14. Choi YJ, Kim HS, Lee SH, Park JS, Nam HS, Kim HJ, et al. Evaluation of peptide nucleic acid array for the detection of hepatitis B virus mutations associated with antiviral resistance. Arch Virol. 2011. 156:1517–1524.

15. Park H, Jang H, Song E, Chang CL, Lee M, Jeong S, et al. Detection and genotyping of Mycobacterium species from clinical isolates and specimens by oligonucleotide array. J Clin Microbiol. 2005. 43:1782–1788.
16. Pellestor F, Paulasova P, Hamamah S. Peptide nucleic acids (PNAs) as diagnostic devices for genetic and cytogenetic analysis. Curr Pharm Des. 2008. 14:2439–2444.

17. Kim YJ, Park MY, Kim SY, Cho SA, Hwang SH, Kim HH, et al. Evaluation of the performances of AdvanSure TB/NTM real time PCR kit for detection of mycobacteria in respiratory specimens. Korean J Lab Med. 2008. 28:34–38.

18. Behr MA, Warren SA, Salamon H, Hopewell PC, Ponce de Leon A, Daley CL, et al. Transmission of Mycobacterium tuberculosis from patients smear-negative for acid-fast bacilli. Lancet. 1999. 353:444–449.
19. Dinnes J, Deeks J, Kunst H, Gibson A, Cummins E, Waugh N, et al. A systematic review of rapid diagnostic tests for the detection of tuberculosis infection. Health Technol Assess. 2007. 11:1–196.

20. Greco S, Girardi E, Navarra A, Saltini C. Current evidence on diagnostic accuracy of commercially based nucleic acid amplification tests for the diagnosis of pulmonary tuberculosis. Thorax. 2006. 61:783–790.

21. Marras TK, Chedore P, Ying AM, Jamieson F. Isolation prevalence of pulmonary non-tuberculous mycobacteria in Ontario, 1997-2003. Thorax. 2007. 62:661–666.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download