Abstract
Pallister-Killian syndrome (PKS) is a rare multisystem disorder characterized by isochromosome 12p and tissue-limited mosaic tetrasomy 12p. In this study, we diagnosed three pediatric patients who were suspicious of having PKS using array-based comparative genomic hybridization (array CGH) and FISH analyses performed on peripheral lymphocytes. Patients 1 and 2 presented with craniofacial dysmorphic features, hypotonia, and a developmental delay. Array CGH revealed two to three copies of 12p in patient 1 and three copies in patient 2. FISH analysis showed trisomy or tetrasomy 12p. Patient 3, who had clinical features comparable to those of patients 1 and 2, was diagnosed by using FISH analysis alone. Here, we report three patients with mosaic tetrasomy 12p. There have been only reported cases diagnosed by chromosome analysis and FISH analysis on skin fibroblast or amniotic fluid. To our knowledge, patient 1 was the first case diagnosed by using array CGH performed on peripheral lymphocytes in Korea.
Pallister–Killian syndrome (PKS) (Online Mendelian Inheritance in Man #601803) is a rare genetic disorder characterized by craniofacial dysmorphisms including a prominent forehead with sparse frontotemporal hair and a wide range of developmental delays, intellectual disability, hypotonia, seizures, skin pigmentation, and other systemic anomalies [1]. PKS is diagnosed by the presence of tetrasomy 12p, which consists of additional short arms of chromosome 12. The additional short arms are usually detected as isochromosomes of 12p with tissue-limited mosaicism [2], which is rarely found in peripheral blood; therefore, the diagnosis usually requires analysis of cultured fibroblasts [3]. Array-based comparative genomic hybridization (array CGH) may enable detection of copy number variations using DNA isolated from an uncultured tissue sample. This method has improved molecular diagnostic detection of clinically significant chromosomal aberrations.
In this study, we identified three patients with mosaic tetrasomy 12p. There have been a few case reports in Korea, in which all patients were detected by chromosome or FISH analysis. To our knowledge, these are the first reported cases of PKS detected by array CGH in Korea. The first two patients were initially identified by array CGH, and the other patient was diagnosed with FISH analysis alone. The genetic study was performed with the written informed consent of the parents. This study was exempted from approval by institutional review board of the hospital.
A 4-yr-old boy presented with a developmental delay with a large hypopigmented macule on his face. Chromosome analysis of peripheral lymphocytes revealed karyotype 46,XY. Array CGH was performed by using the Affymetrix Cytogenetics 2.7M Array (Affymetrix; Santa Clara, CA, USA) with DNA extracted from peripheral lymphocytes, which identified two to three copies of chromosome 12p. This finding was suggestive of chimerism between trisomy 12p and tetrasomy 12p (Fig. 1A). FISH analysis by means of ETV6 (TEL)/RUNX1 (AML1) Extra signal dual color probe (Abbott Molecular Inc., Des Plaines, IL, USA) was performed on cultured peripheral lymphocytes according to the manufacturer's instructions. This assay revealed that 2.3% and 3.5% of the cells were positive for trisomy 12p and tetrasomy 12p, respectively (Fig. 1B).
A 4-month-old female infant was referred due to multiple congenital anomalies with a hypopigmented macule. Chromosomal analysis revealed 46,XX. To confirm the suspected clinical features, FISH analysis was conducted to identify possible additional copies of chromosome 12. Approximately 11% of the cells showed tetrasomy 12p (Fig. 1B).
PKS is a rare chromosomal disorder with characteristic features that change with the patient's age [4]. Craniofacial abnormalities are prevalent in most patients. Nearly 90% of patients have ophthalmologic abnormalities, and over 75% have a hypopigmented skin lesion and hearing loss [5]. Our cases had the common dysmorphic facial features such as frontal bossing, a depressed nasal bridge, frontotemporal balding, and abnormal skin pigmentation. The patients presented here are similar to those reported in other studies.
Different tissues are known to show variable presence or absence of isochromosome 12p, i(12p) [6]. The level of mosaicism varies depending on the gestational age or tissue-specific mosaicism [6]. The patients described in this report initially had normal chromosomal results on cultured peripheral lymphocytes.
We identified three patients with mosaic tetrasomy 12p. To our knowledge, these are the first reported cases of PKS detected by array CGH in Korea. The first two patients were initially identified by array CGH, and the other patient was diagnosed with FISH analysis alone. It is likely that FISH analysis is sufficient for identifying PKS because FISH confirmed the presence of mosaicism in the three patients presented here. In the prenatal and perinatal periods, however, the clinical characteristics of PKS are not prominent. Therefore, conventional chromosomal analyses may yield normal results at these ages, necessitating array CGH for definitive diagnosis.
Mosaicism in the supernumerary metacentric isochromosome 12p is often detected in skin fibroblasts although the karyotype of cultured lymphocytes appears normal. There seems to be a higher success rate of detecting mosaicism in cultured skin fibroblasts than in peripheral blood because abnormal cells have a lower attrition rate than T lymphocytes do [67]. Furthermore, cell culture may select against abnormal cells. Phytohemagglutinin is often used in chromosomal analysis to stimulate T lymphocytes to divide. If abnormal cells are less competitive than are normal cells in stem cell populations, then they may be underrepresented in stimulated cultures [89]. Therefore, cells with isochromosome 12p are likely to be selected against during cell culture [10]. Ballif et al [11] found that the percentage of abnormal cells among phytohemagglutinin-stimulated cells is lower than that observed in uncultured cells when FISH is used for analysis. This finding suggests that there is a chromosome-or chromosomal-region-specific growth bias in cell culture. Furthermore, a rapid decline of abnormal clones [with i(12p)] was discovered during amniocyte subculture [12], and the death of these abnormal cells has been suggested as the cause of tissue-limited mosaicism in PKS [13]. Low-level mosaicism of chromosomal abnormalities with clinical significance can be overlooked when masked by a high proportion of normal cells or by culturing bias that is necessary for conventional cytogenetic testing [11]. Wilkens et al [5] reviewed the existing literature and stated that peripheral lymphocytes show a higher percentage of cells with a healthy karyotype and a lower percentage of i(12p) mosaicism. The correct diagnosis has probably been overlooked in a number of patients because skin fibroblasts were not routinely included in karyotyping in the past [1415].
Array CGH is useful for patients who have suspected clinical and/or constitutional abnormalities. This method can rapidly screen genomes at an unprecedented resolution and has dramatically improved the molecular diagnostic detection of clinically significant chromosomal aberrations [1617]. Array CGH has several advantages over FISH. The former using extracted DNA, does not require cell culture, which can introduce culturing bias [10]. In addition, cells in all cell cycle phases can be analyzed simultaneously [18]. However, array CGH of unstimulated peripheral lymphocytes has failed to detect all cases of PKS because it can detect mosaic abnormal cells at prevalence not lower than 10-20% [1011]. Analysis of skin fibroblasts [19] or buccal smears [20] has been proposed as a diagnostic gold standard, but this notion is still controversial. Hodge et al [19] proposed that a skin biopsy sample should continue to be the diagnostic gold standard for PKS because of the variable and insufficient quality of buccal smears. Nonetheless, Cobben et al [20] showed that in patients with suspected PKS, array CGH or FISH analysis of buccal smear cells seems to be the first diagnostic choice because these two methods are reliable, rapid, and noninvasive. Cobben et al [20] recommended a skin biopsy only if buccal-smear analysis cannot detect i(12p).
PKS is a rare multisystem disorder that is difficult to diagnose prenatally and in early infancy. The diagnosis of PKS can be made by physical examination if characteristic dysmorphic features and global developmental delay are present. Array CGH can be used to diagnose PKS in early infancy and during the prenatal period. Clinicians can also employ FISH analysis to diagnose this disorder in patients with visible PKS features.
Acknowledgments
We thank the patients and their parents for their participation in this study. We are also grateful to the staff of the Department of Cytogenetics at Samsung Medical Center for their excellent technical assistance.
References
1. Pallister PD, Meisner LF, Elejalde BR, Francke U, Herrmann J, Spranger J, et al. The pallister mosaic syndrome. Birth Defects Orig Artic Ser. 1977; 13:103–110.
2. Peltomäki P, Knuutila S, Ritvanen A, Kaitila I, de la Chapelle A. Pallister-Killian syndrome: cytogenetic and molecular studies. Clin Genet. 1987; 31:399–405. PMID: 2887316.
3. Wenger SL, Boone LY, Steele MW. Mosaicism in Pallister i(12p) syndrome. Am J Med Genet. 1990; 35:523–525. PMID: 2333883.
4. Schinzel A. Tetrasomy 12p (Pallister-Killian syndrome). J Med Genet. 1991; 28:122–125. PMID: 2002482.
5. Wilkens A, Liu H, Park K, Campbell LB, Jackson M, Kostanecka A, et al. Novel clinical manifestations in Pallister-Killian syndrome: comprehensive evaluation of 59 affected individuals and review of previously reported cases. Am J Med Genet A. 2012; 158A:3002–3017. PMID: 23169767.
6. Priest JH, Rust JM, Fernhoff PM. Tissue specificity and stability of mosaicism in Pallister-Killian +i(12p) syndrome: relevance for prenatal diagnosis. Am J Med Genet. 1992; 42:820–824. PMID: 1554021.
7. Reeser SL, Wenger SL. Failure of PHA-stimulated i(12p) lymphocytes to divide in Pallister-Killian syndrome. Am J Med Genet. 1992; 42:815–819. PMID: 1554020.
8. Pagon RA, Hall JG, Davenport SL, Aase J, Norwood TH, Hoehn HW. Abnormal skin fibroblast cytogenetics in four dysmorphic patients with normal lymphocyte chromosomes. Am J Hum Genet. 1979; 31:54–61. PMID: 155399.
9. Kulharya AS, Lovell CM, Flannery DB. Unusual mosaic karyotype resulting from adjacent 1 segregation of t(11;22): importance of performing skin fibroblast karyotype in patients with unexplained multiple congenital anomalies. Am J Med Genet. 2002; 113:367–370. PMID: 12457409.
10. Theisen A, Rosenfeld JA, Farrell SA, Harris CJ, Wetzel HH, Torchia BA, et al. aCGH detects partial tetrasomy of 12p in blood from Pallister-Killian syndrome cases without invasive skin biopsy. Am J Med Genet A. 2009; 149A:914–918. PMID: 19353629.
11. Ballif BC, Rorem EA, Sundin K, Lincicum M, Gaskin S, Coppinger J, et al. Detection of low-level mosaicism by array CGH in routine diagnostic specimens. Am J Med Genet A. 2006; 140:2757–2767. PMID: 17103431.
12. Polityko AD, Goncharova E, Shamgina L, Drozdovskaja N, Podleschuk L, Abramchik E, et al. Pallister-Killian syndrome: rapid decrease of isochromosome 12p frequency during amniocyte subculturing. Conclusion for strategy of prenatal cytogenetic diagnostics. J Histochem Cytochem. 2005; 53:361–364. PMID: 15750020.
13. Tang W, Wenger SL. Cell death as a possible mechanism for tissue limited mosaicism in Pallister-Killian syndrome. J Assoc Genet Technol. 2005; 31:168–169. PMID: 16354943.
14. Hall BD. Mosaic tetrasomy 21 is mosaic tetrasomy 12p some of the time. Clin Genet. 1985; 27:284–285. PMID: 3987079.
15. Lopes V, Mak E, Wyatt PR. Prenatal diagnosis of tetrasomy 21. Prenat Diagn. 1985; 5:233–235. PMID: 3161017.
16. Shaffer LG, Kashork CD, Saleki R, Rorem E, Sundin K, Ballif BC, et al. Targeted genomic microarray analysis for identification of chromosome abnormalities in 1500 consecutive clinical cases. J Pediatr. 2006; 149:98–102. PMID: 16860135.
17. Bejjani BA, Theisen AP, Ballif BC, Shaffer LG. Array-based comparative genomic hybridization in clinical diagnosis. Expert Rev Mol Diagn. 2005; 5:421–429. PMID: 15934818.
18. Spinner NB, Conlin LK. Mosaicism and clinical genetics. Am J Med Genet C Semin Med Genet. 2014; 166C:397–405. PMID: 25424979.
19. Hodge JC, Hulshizer RL, Seger P, St Antoine A, Bair J, Kirmani S. Array CGH on unstimulated blood does not detect all cases of Pallister-Killian syndrome: a skin biopsy should remain the diagnostic gold standard. Am J Med Genet A. 2012; 158A:669–673. PMID: 22315202.
20. Cobben JM, Engelen M, Polstra A. Array CGH on unstimulated blood does not detect all cases of Pallister-Killian syndrome: buccal smear analysis should remain the diagnostic procedure of first choice. Am J Med Genet A. 2013; 161A:1517–1519. PMID: 23613446.
Fig. 1
Genomic analysis. (A) Array CGH analysis of chromosome 12 performed on DNA from blood lymphocytes. Log 2 ratio data are presented according to the position in the human genome (NCBI build 37/hg19). Patient 1 showed two to three copies of 12p13.33-p11.21 (arr 12p13.33p11.21×2-3). Patient 2 had a duplication of the entire short arm of chromosome 12, 12p13.3-p11.1 (arr 12p13.3p11.1×3). (B) FISH results from three patients. FISH analysis revealed four copies in addition to the three copies of TEL (green) on 12p13 in patient 1 and four copies of TEL (green) on 12p13 in patients 2 and 3. The red signal indicates AML1 (21q22).
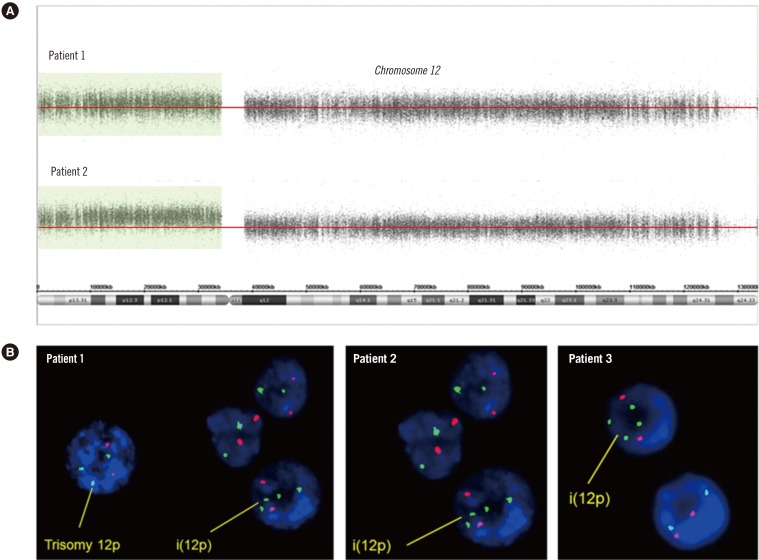
Table 1
Patient phenotypes as compared to a prior description of Pallister-Killian syndrome [4]
Table 2
Summary of cytogenetic and FISH analyses of peripheral lymphocytes and array CGH of peripheral-lymphocyte DNA





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download