Abstract
Objectives
There are 3 subtypes of natriuretic peptide (NP) receptors: type A natriuretic peptide receptor (NPRA), NPRB, and NPRC. The NPRA gene polymorphism, consisting of substition of methionine (ATG) to isoleucine (ATC) at nucleotide 1023 (M341I) of exon 3 was revealed to be associated with increased risk for essential hypertension (EH) in Japanese people. The purpose of this study is to investigate association between EH and the M341I polymorphism in the NPRA gene in Korea.
Methods
Eighty patients in whom type B natriuretic peptide (BNP) was measured were enrolled in this study. 66 patients had EH and 14 patients did not. The polymorphism of M341I was evaluated by multiplex genotyping polymerase chain reaction and by sequencing analysis.
Results
The overall distribution of alleles was not significantly different between the control and EH groups. However, the C/C homozygous genotype was found only in the EH group. In the EH group, patient carrying the C/C homozygous genotype had the trend of having higher systolic and diastolic BP levels regardless of the previous treatment, even though other laboratory markers including BNP levels had no significant differences according to the genotypes.
Natriuretic peptide (NP) effects are mediated by binding to cell surface receptors. There are 3 subtypes of NP receptors: type A natriuretic peptide receptor (NPRA) [1], type B natriuretic peptide receptor (NPRB) [2], and type C natriuretic peptide receptor (NPRC) [3]. All 3 types affect cellular second messenger activity. NPRA and NPRB are membrane-bound guanylate cyclase and their activation increases cGMP levels [4]. Activation of NPRC results in inhibition of adenylyl cyclase activity [5]. Human NPRA (hNPRA) has high structural homology with human NPRB (hNPRB), and contains a highly conserved guanylyl cyclase domain [3]. Type A natriuretic peptide (ANP) and Type B natriuretic peptide (BNP) bind primarily to NPRA, which is found in the vasculature, causing vasodilation and inhibition of vascular smooth muscle cell proliferation [6]. Therefore, absence or decreased activity of this receptor may result in hypertension or other cardiovascular diseases [1-6].
Several studies have discovered mutations in the hNPRA gene which may be associated with decreased activity of the hNPRA [7-14].
The organization of the hNPRA gene was recently described by Takahashi et al. [15], Nakayama et al. [16]. They also discovered a novel missense mutation M341I in the hNPRA gene which is consisting of a methionine (ATG) to isoleucine (ATC) substitution at nucleotide 1023 in exon 3. The M341I mutation was revealed to be associated with increased risk for essential hypertension (EH) in Japanese people [9].
However, this mutation has not been investigated in other countries or in other ethnics. So, the purpose of our study is to investigate the polymorphism of the M341I and the association between the M31I polymorphism and EH in Korea.
Eighty patients in whom BNP was measured were enrolled in this study. The mean age of the patient population was 62 years, with 41 male and 39 female participants. Participants were patients attending the out-patient clinics of Ewha Womans University Dongdaemun Hospital in Seoul, Korea, from January to May 2005. All participants gave informed consent. The study complies with the Declaration of Helsinki and was approved by the local ethics committee of the Ewha Womans University Dongdaemun Hospital.
EH was defined by the administration of antihypertensive agents, a systolic blood pressure greater than 140 mmHg or a diastolic blood pressure greater than 90 mmHg on three occasions during the 2 months following the first medical examination. 66 patients had EH and 14 patients did not.
BNP was measured in plasma using a point-of-care immunoassay from Biosite Triage BNP (Biosite Diagnostics, San Diego, CA, USA). C-reactive protein (CRP), lactate dehydrogenase (LDH), uric acid (UA), total cholesterol, high density lipoprotein-cholesterol (HDL-C), low density lipoprotein-cholesterol (LDL-C), triglyceride (TG) were measured using the Hitachi 7600-110 analyzer (Hitachi, Tokyo, Japan) and Wako reagents (Wako Pure Chemical Industries, Tokyo, Japan). Homocysteine (Hcy) was measured in plasma using the AxSYM automated analyzer (Abbott Laboratories, Abbott Park, IL, USA).
The polymorphism of M341I was evaluated by multiplex genotyping polymerase chain reaction (PCR) using dual specificity oligonucleotide (DSO) primers from Seegene (Seegene Institute of Life Sciences, Seoul, Korea).
Genomic DNA was extracted from whole blood leukocytes by a standard method using genomic DNA isolation kit (Genomic blood spin mini kit, Nucleogen Biotechnology, Siheung, Korea). Identification of the G to C transition at nucleotide 1023 was performed by polymerase chain reaction amplification of a 672 bp segment containing nucleotide 1023. Four DSO primers were used in genotyping of the hNPRA gene. NPR-F primer was 5'TCCAGTTTGACCTTGAGCIIIIIAGAGACGC3', NPR-MR primer was 5'GGACCCCTTCTTACCAGIIIIICCTCGATG3', NPR-WF primer was 5'GGCCTATGAGCAGTTCAAIIIIICCATGGAG3', NPR-R primer was 5'AAGGATGCTGGGATGGIIIIIACCTGTAG3'. Target regions of NPR-F, NPR-MR, NPR-WF, NPR-R were DNA sequences of 3019-3049, 3242-3271, 3218-3248, and 3662-3690, respectively. NPR-F/NPR-R primer set was for the 672 bp fragments, NPR-F/NPR-MR primer set was for the 253 bp fragments, and NPR-WF/NPR-R primer set was for the 473 bp fragments. Amplification was performed in the DNA thermal cycler (Perkin Elmer Co., Norwalk, CT, USA) with an initial denaturing step at 94℃ for 15 minutes, followed by 35 cycles of denaturing at 94℃ for 30 seconds, annealing at 65℃ for 30 seconds, and extension at 72℃ for 1 minute. The fragment was amplified in a reaction volume of 20 µL containing 20-30 ng genomic DNA, 4 µL of 5X primer mixture (final 0.25 µM each), 10 µL of 2X Master Mix. The amplified PCR products were separated in 2% agarose gel stained with ethidium bromide. Wild homozygote showed 2 bands which were 672 bp fragment and 473 bp fragment. Mutant homozygote showed 2 bands which were 672 bp fragment and 253 bp fragment. Mutant Heterozygote showed 3 bands which were 672 bp fragment, 473 bp fragment, and 253 bp fragment.
We confirmed the function of above primers using PCR fragments including wild homozygote, mutant homozygote and mutant heterozygote of the missense mutation of the exon 3 (M341I) from original Japanese study [9].
The target PCR products within the agarose gel were purified for sequencing using the QIAquick PCR purification kit (Quiagen, Valencia, CA, USA). The purified PCR products were sequenced in ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Statistical analyses were performed with χ2 analysis, Student's t test, Mann-Whitney U test using SPSS version 11.0 (SPSS Inc., Chicago, IL, USA).
The PCR analysis confirming the function of DSO primers are shown in Fig. 1. The G/C mutant hetero zygote shows three bands of 672, 473, and 253 bps. The G/G wild homozygote shows two bands of 672 and 473 bps. The C/C mutant homozygotes show two bands of 672 and 253 bps. The sequencing analysis of the PCR products are also shown (Fig. 1). The PCR analysis and the sequencing analysis of the patient samples are shown (Fig. 2).
The characteristics of the study participants are shown in Table 1. UA was higher in the EH group than in the control subjects (P=0.004). Although other laboratory parameters showed no significantly different concentrations between in control subjects and in the EH group, there were trends of higher levels of laboratory parameters including BNP, CRP, Hcy, and LDH in the EH group compared with those of the control subjects (Table 1).
The results of genotyping of M341I polymorphism are shown in Table 2. In the control subjects and in the EH group, the prevalence of the G/G wild homozygote was 92.9% (13/14), 75.8% (50/66), the G/C heterozygous genotype was 7.1% (1/14), 22.7% (15/66), respectively. The C/C homozygous genotype was found only in the EH group. These data did not agree with predicted Hardy-Weinberg equilibrium values (χ2=54.53, P=0.000). Although not statistically significant, the C allele was found more frequently in the EH group than in the control subjects (Table 2).
In the EH group, patient carrying the C/C homozygous genotype had the trend of having higher systolic and diastolic BP levels regardless of the previous treatment, even though other laboratory markers including BNP levels had no significant differences according to the genotypes (Table 3).
Several mutations in the NPRA gene were discovered [7-14]. An 8-bp deletion at the transcription factor AP2 site in the untranslated region of exon 1 was discovered by analyzing the 5'-flanking region of the NPRA gene. This deletion in the NPRA gene reduces receptor activity and may confer increased susceptibility to developing EH or left ventricular hypertrophy in Japanese individuals [7].
D893A mutation in the cyclase catalytic region (HCAT) of the NPRA was identified by Thompson and Garbers [8]. In the presence of this mutation, the cGMP levels in response to ANP in vitro was inhibited by 85% of the full-length guanylyl cyclase-A activity of the NPRA [8].
A novel missense mutation, M341I, consisting of a methionine (ATG) to isoleucine (ATC) substitution at nucleotide 1023 in exon 3 of the NPRA gene was discovered in Japanese individuals. The overall distribution of alleles was not significantly different between the control and EH groups. However, the C/C homozygous genotype was found only in the EH group. The authors concluded that the significance of homozygous M341I mutation in exon 3 is worth investigating for its possible association with EH [9].
Another study concludes that the M341I missense mutation is associated with risk for MI and may be a genetic marker of MI in Japanese people [10].
A novel insertion/deletion variant at position 15,129 in the 3'-untranslated region (3'-UTR) of NPRA receptor gene, the deletion 15129 was also discovered. The "deletion 15129" variant might participate in the functional impairment of natriuretic peptide system defining an increased genetic susceptibility to hypertension [11].
Another genetic susceptibility of NPRA to EH and cardiac hypertrophy was determined. The authors concluded that common NPRA alleles can alter expression of the gene as much as two-fold and could therefore significantly affect genetic risks for EH and cardiac hypertrophy in humans [12].
Transgenic mice overexpressing the NPRA gene had significantly lower blood pressure than wild type mice. The observed NPRA gene-dose effect on blood pressure variation suggests that, in humans, differences in the expression of the NPRA gene and protein are also reflected in differences in blood pressure. Therefore, the gene-dose effect is also important in providing the rationale for the genetic analysis of the NPRA gene in essential hypertension [13].
But, in a postmyocardial infarction cohort from New Zealand, there was no NPRA gene functional deletion allele which had been discovered in Japanese individuals [14]. These findings suggest that the prevalences of these genetic variations might be various according to various diseases or ethnics.
In Korean individuals, we identified the polymorphism of M341I of the NPRA gene. We detected the G/C heterozygous genotype in both groups of control subjects and the EH group. However, the C/C homozygous genotype was found only in the EH group. In the EH group, patient carrying the C/C homozygous genotype had the trend of having higher systolic and diastolic BP levels regardless of the previous treatment, even though other laboratory markers had no significant differences according to the genotypes. These findings uphold the previous study [9] suggesting that the C allele may influence the function of NPRA in C/C homozygous subjects, but not in G/C heterozygosity. Several genetic receptor abnormalities have revealed autosomal recessive conditions that reside predominantly in the homozygous state of the mutation [9].
In this study, patient carrying the C/C homozygous genotype had lower BNP concentrations compared with those of the other patients carrying the G/G or the G/C genotype. This finding is not consistent with the previous study [9] in which the ratio of plasma BNP/mean blood pressure of the C/C genotype was significantly higher than that of the G/G genotype or the G/C genotype. This study suggests that NPRA's actions are not only the receptor functions of the BNP, but also the other biological activities associated with developing the EH. This study suggests that the C/C genotype might develop EH before the BNP concentration is elevated with different mechanism. Furthermore, the patient carrying the C/C homozygous genotype might also have the mutation in the BNP gene which could down-regulate the BNP gene transcription resulting in decreased BNP concentration. But, there are another study of NPRA gene polymorphism in which the BNP levels showed no statistically significant difference according to the NPRA polymorphism in the EH patients, which strongly upholds our result [17].
EH might be caused by multiple factors including various genetic mutations beside of the M341I polymorphism. Further studies would be necessary to detect not only the M341I polymorphism but also the other mutations in large numbers of EH patients.
Even though this study has major limitation of small numbers of control group and EH patient group, this has the concise meaning as the first study about the M341I polymorphism in the NPRA gene in Korea. This is the first study using the multiplex genotyping PCR with DSO primers in identifying the M341I polymorphism. We detected the C/C homozygous genotype only in the EH group which suggests the association of M341I polymorphism with the EH in Korea.
Figures and Tables
Fig. 1
(A) The PCR analysis confirming the function of dual specificity oligonucleotide (DSO) primers. The G/C mutant heterozygote in lane 1 shows three bands of 672, 473, and 253 bps. The G/G wild homozygote in lane 2 shows two bands of 672 and 473 bps. The C/C mutant homozygotes in lane 3 and in lane 4 show two bands of 672 and 253 bps (The DNAs are provided by Nakayama et al. [9]). (B) The sequencing analysis of the PCR products.
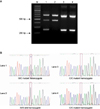
Fig. 2
(A) The PCR analysis of the patient samples. Patient No. 2 shows C/C mutant homozygote. Patient No. 1, 5 show G/C mutant heterozygotes. Others show G/G wild homozygotes. (B) The sequencing analysis of the PCR products of the patient samples.

References
1. Fuller F, Porter JG, Arfsten AE, Miller J, Schilling JW, Scarborough RM, et al. Atrial natriuretic peptide clearance receptor: complete sequence and functional expression of cDNA clones. J Biol Chem. 1988. 263:9395–9401.
2. Maack T, Camargo MJ, Kleinert HD, Laragh JH, Atlas SA. Atrial natriuretic factor: structure and functional properties. Kidney Int. 1985. 27:607–615.
3. Yuen PS, Garbers DL. Guanylyl cyclase-linked receptors. Annu Rev Neurosci. 1992. 15:193–225.
4. Meloche S, McNicoll N, Liu B, Ong H, De Léan A. Atrial natriuretic factor R1 receptor from bovine adrenal zona glomerulosa: purification, characterization, and modulation by amiloride. Biochemistry. 1988. 27:8151–8158.
5. Schenk DB, Phelps MN, Porter JG, Fuller F, Cordell B, Lewicki JA. Purification and subunit composition of atrial natriuretic peptide receptor. Proc Natl Acad Sci U S A. 1987. 84:1521–1525.
6. Bennett BD, Bennett GL, Vitangcol RV, Jewett JR, Burnier J, Henzel W, et al. Extracellular domain-IgG fusion proteins for three human natriuretic peptide receptors: hormone pharmacology and application to solid phase screening of synthetic peptide antisera. J Biol Chem. 1991. 266:23060–23067.
7. Nakayama T, Soma M, Takahashi Y, Rehemudula D, Kanmatsuse K, Furuya K. Functional deletion mutation of the 5'-flanking region of type A human natriuretic peptide receptor gene and its association with essential hypertension and left ventricular hypertrophy in the Japanese. Circ Res. 2000. 86:841–845.
8. Thompson DK, Garbers DL. Dominant negative mutations of the guanylyl cyclase-A receptor: extracellular domain deletion and catalytic domain point mutations. J Biol Chem. 1995. 270:425–430.
9. Nakayama T, Soma M, Mizutani Y, Xinjuan X, Honye J, Kaneko Y, et al. A novel missense mutation of exon 3 in the type A human natriuretic peptide receptor gene: possible association with essential hypertension. Hypertens Res. 2002. 25:395–401.
10. Nakayama T, Soma M, Saito S, Honye J, Sato M, Aoi N, et al. Missense mutation of exon 3 in the type A human natriuretic peptide receptor gene is associated with myocardial infarction. Med Sci Monit. 2003. 9:CR505–CR510.
11. Lucarelli K, Iacoviello M, Dessì-Fulgheri P, Sarzani R, Romito R, Sorrentino S, et al. Natriuretic peptides and essential arterial hypertension. Ital Heart J Suppl. 2002. 3:1085–1091.
12. Knowles JW, Erickson LM, Guy VK, Sigel CS, Wilder JC, Maeda N. Common variations in noncoding regions of the human natriuretic peptide receptor A gene have quantitative effects. Hum Genet. 2003. 112:62–70.
13. Nakayama T. The genetic contribution of the natriuretic peptide system to cardiovascular diseases. Endocr J. 2005. 52:11–21.
14. Palmer BR, Frampton CM, Richards AM, Cameron VA, Nakayama T. Absence of a NPR-A gene functional deletion allele in a postmyocardial infarction cohort from New Zealand. Circ Res. 2004. 94:e86.
15. Takahashi Y, Nakayama T, Soma M, Izumi Y, Kanmatsuse K. Organization of the human natriuretic peptide receptor A gene. Biochem Biophys Res Commun. 1998. 246:736–739.
16. Nakayama T, Soma M, Takahashi Y, Rehemudula D, Sato M, Uwabo J, et al. Nucleotide sequence of the 5'-flanking region of the type A human natriuretic peptide receptor gene and association analysis using a novel microsatellite in essential hypertension. Am J Hypertens. 1999. 12:1144–1148.
17. Rubattu S, Bigatti G, Evangelista A, Lanzani C, Stanzione R, Zagato L, et al. Association of atrial natriuretic peptide and type a natriuretic peptide receptor gene polymorphisms with left ventricular mass in human essential hypertension. J Am Coll Cardiol. 2006. 48:499–505.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download