INTRODUCTION
Diabetes mellitus (DM), which is the sixth leading cause of death in Korea, has a prevalence rate of 13.9% in adults older than 30 years and that of 27.6% in those aged over 65. As the prevalence of diabetes in older individuals is expected to increase further due to population aging, necessary preparation is warranted [
1].
Older adults with diabetes are more likely to experience complications and have a higher risk of cardiovascular disease than younger adults because blood sugar control becomes more difficult due to the physiological changes associated with aging [
2]. Diabetes is a chronic disease that requires lifelong management because it is difficult to cure; moreover, to reduce the prevalence of diabetes, its early prevention is extremely important. Smoking, lack of physical activity, and poor eating habits have been reported as major risk factors for diabetes [
3]. As dietary habits are less challenging to control than other factors, establishing an optimal nutrient intake is imperative for diabetes prevention and management.
Despite strong presumptive evidence of relationships between certain diseases and particular nutrients, studies have often demonstrated negligible or no associations between them [
4]. Data analyzed using general statistical techniques including linear regression are often difficult to interpret correctly when the sample size is small, the functional relationship is unknown, or the relationship among several factors is complex and highly correlated; furthermore, identifying a specific inherent type or influencing factor is not easy. The neural network proposed to address this challenge is an artificial intelligence data analysis method that identifies patterns or knowledge hidden in large-scale data and showed better performance against linear regression in predicting diabetes [
5]. In the early days, it was merely a simple logical operator; however, with the rapid advancement of computer calculation speed, its application field has gradually widened, and it has actively been used in medical systems for diagnosis and prediction of diseases such as hypertension [
6], hyperlipidemia [
7], and cardiac arrhythmia [
8].
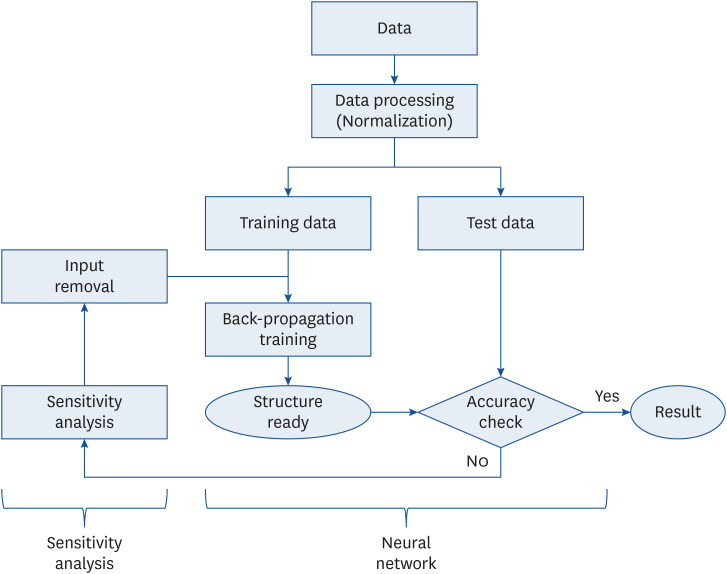
When configuring a neural network system, increasing the number of input factors complicates the system’s structure, thus amplifying the amount of computation and consuming considerable time. After simplifying the system and enhancing its performance by excluding input factors with a relatively low contribution, identifying factors essential for generating an efficient system becomes expedient [
91011].
The disadvantage of the neural network system is that only the output value can be derived, and the extent to which the input variable contributes to this output cannot be determined. By incorporating sensitivity analysis into the neural network system, the relative contribution of each participating input factor can be expressed as a quantified number [
121314]. Therefore, it is possible to develop an optimized neural network system that reveals the maximum predictive accuracy when the minimum input factors necessary for constructing a neural network are determined through an iterative process of sequentially removing minimally contributing factors and monitoring the prediction-rate trend. By so doing, the predictive importance of each input factor can be determined. To date, most studies on diabetes prediction using neural networks have evidently employed non-nutrient input factors, such as body mass index (BMI), diastolic blood pressure, stress, smoking, and age [
151617], and few studies have constructed neural network systems based on nutrient factors. In particular, few cases exist in which sensitivity analysis has been applied to improve the prediction rate when constructing a system.
Therefore, this study established a nutrient intake-based diabetes-prediction system for the older population using neural network sensitivity analysis. The contribution of each nutrient to diabetes prediction was determined to pave way for future nutrient intake-based diabetes predictions in older individuals. This study’s findings will prove useful as fundamental data for customized nutrition education based on the identified nutrients.
DISCUSSION
Older adults with diabetes encounter multifaceted challenges, such as economic, physical, and physiological functional problems; thus, managing their blood sugar levels is difficult, and their quality of life deteriorates. Therefore, appropriate customized management is required. On this premise, investigating the nutritional characteristics related to diabetes prevalence in the older population in Korea is considerably significant as it may enable the customized management of older patients with diabetes.
To date, studies on diabetes prediction using neural network systems have predominantly focused on adults, and few studies have paid attention to the older population. In addition, the input factors used to establish diabetes-prediction systems have mainly been BMI, blood pressure, stress, and smoking, and since few studies have used nutrients, obtaining information on the contribution of nutrients to diabetes prediction proves difficult.
This study conducted a neural network sensitivity analysis using nutrient factors to 1) establish an efficient, nutrient-based diabetes-determination neural network system for older KNHANES participants and 2) identify nutrients strongly related to diabetes onset to prevent diabetes. Its findings were intended to be used as fundamental data for customized nutrition education.
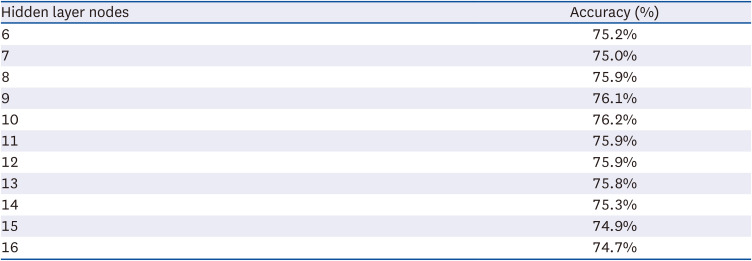
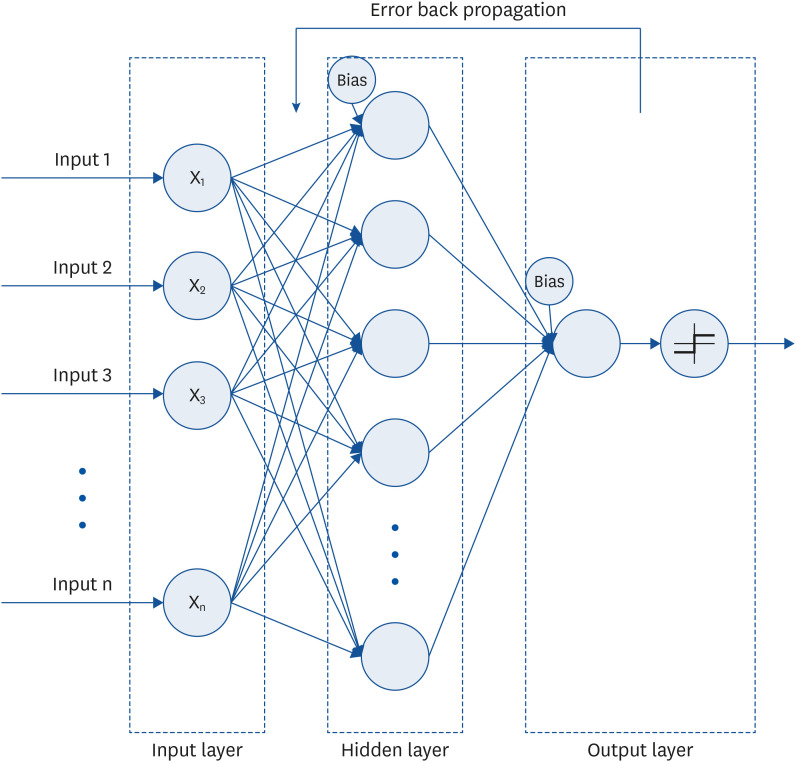
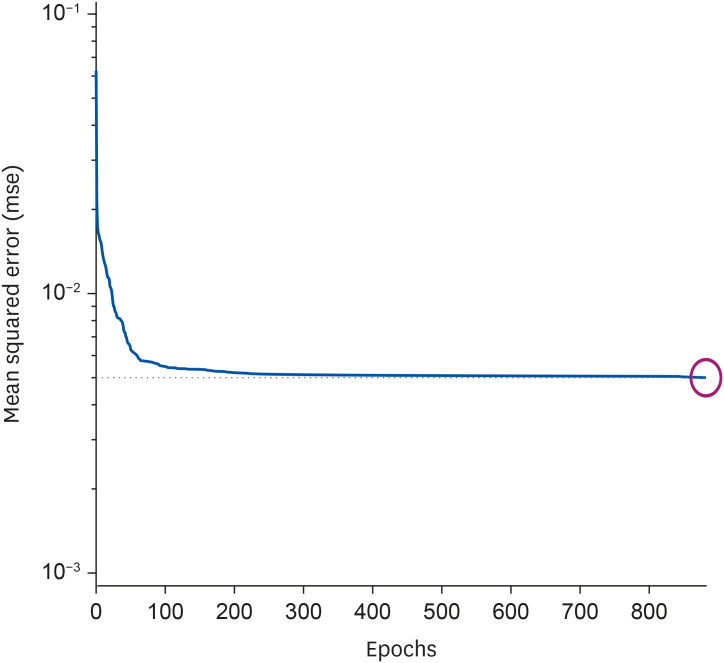
To efficiently configure a neural network’s structure, minimizing the number of constituent neurons (nodes) for each input, hidden, or output layer is necessary. In this study, 16 nodes were set to accommodate 16 nutrients in the input layer, and one node was set to predict diabetes in the output layer. Since predictive accuracy varies depending on the number of nodes in the hidden layer, 10 nodes with the highest predictive accuracy were identified via repeated experiments. Depending on the research conditions, the number of nodes in the neural network’s hidden layer was found to differ. A previous study that constructed a neural network for predicting diabetes in adults in the United States used 12 nodes [
16] in the hidden layer, whereas another that constructed a neural network for predicting blood sugar in Korean adults used 7 [
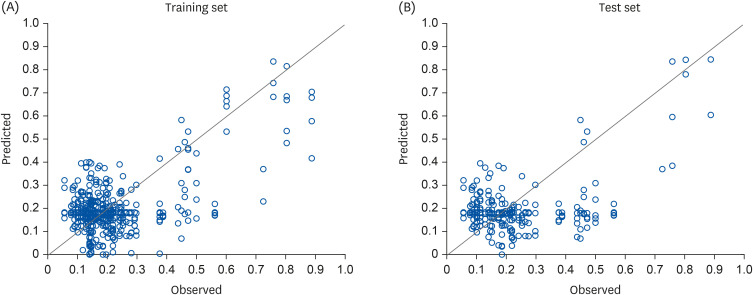
25]. The accuracy of the training process using training data was 84.1%, while that of the test phase using test data was 76.2%. In general, the predictive accuracy of the testing stage tends to be lower than that of the training stage [
26].
In this study, a neural network system was developed based on 16 nutrients, namely, energy, carbohydrates, protein, fat, sugar, cholesterol, vitamin A, thiamine, riboflavin, niacin, vitamin C, calcium, phosphorus, sodium, potassium, and iron, which have not been used as input factors in previous studies. The diabetes predictive accuracy was 76.2%, which is not lower than that yielded by a non-nutrient-based neural network that was attempted in previous studies.
Güldoğan
et al. [
27] conducted a study on the establishment of a diabetes-prediction neural network system using the American Pima Indian female database, which is characterized by a high diabetes incidence rate. They used eight input factors as number of pregnancies, plasma glucose concentration, diastolic blood pressure, triceps skinfold thickness, serum insulin, BMI, diabetes pedigree function, and age and achieved predictive accuracy of 78.1%. Ryu
et al. [
28] constructed a diabetes-prediction neural network using KNHANES data and yielded an accuracy of 80.0% using seven input factors: age, gender, hypertension, family history of diabetes, smoking status, BMI, and waist circumference. In a study by Agliata
et al. [
16], a diabetes-prediction neural network system was generated using US National Health and Nutrition Examination Survey data, and the following nine input factors were used: sex, age, high-density lipoprotein (HDL) cholesterol, glucose, systolic blood pressure, diastolic blood pressure, triglycerides, weight, and BMI. Applying these factors yielded a relatively high accuracy of 86%. In contrast, Liu
et al. [
17] constructed a neural network system for predicting diabetes in Chinese older adults aged over 65 years using the following eight input factors: education, BMI, waist circumference, fasting plasma glucose, total cholesterol, triglyceride, HDL-cholesterol, and alanine aminotransferase and achieved a low predictive accuracy of 60.7%.
In a neural network system, configuring input factors with appropriate variables is directly related to improving the training ability of the neural network and securing excellent predictive accuracy. In the case of a model involving numerous input factors, including a factor with a negligible contribution or weak correlation potentially decreases the model’s predictive accuracy; therefore, optimization via a sensitivity analysis technique is required. In addition, the general neural network structure thus far merely shows the resulting predictive accuracy based on input factors; however, it is limited in that it cannot determine how each input factor contributed to the predicted outcome. However, the sensitivity index resulting from neural network sensitivity analysis may indicate the relative importance of each input factor in predicting the output.
Among the various sensitivity analysis methods available, this study selected the perturbation method [
29], which determines the importance of an input factor by monitoring output-factor changes in response to input-variable changes. Compared with other sensitivity analysis methods, such as the stepwise elimination method [
29], which removes input factors sequentially, reconstructs the neural network through an additional training process, and subsequently compares predictive accuracies to determine the importance of input factors, the perturbation method is simpler with fewer calculations and no additional training process, thereby being widely used due to its efficiency and simplicity.
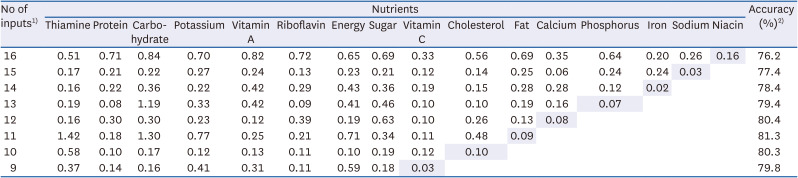
In the neural network system involving 16 nutrients as input factors, the diabetes-prediction accuracy was 76.2%. Through sensitivity analysis, iron, phosphorus, calcium, fat, and cholesterol, which are nutrients of relatively low importance with low sensitivity index values, were sequentially excluded. Consequently, the predictive accuracy peaked at 81.3% with the updated neural network system of 11 input nutrient combinations, and this system was confirmed as the final structure. The 11 input factors of the confirmed optimal neural network structure were energy, carbohydrates, protein, fat, sugar, cholesterol, vitamin A, thiamin, riboflavin, vitamin C, and potassium. In particular, thiamin, carbohydrates, potassium, and energy were found to be relatively more important in predicting diabetes with higher sensitivity index values. Nutrients of high importance in predicting diabetes are not directly related to the mechanism of diabetes onset; nonetheless, they are probably related to diabetes onset to a certain extent. Therefore, in future, attention should be focused on dietary education in diabetes prevention.
Sensitivity analysis enabled a reduction in nutrient input factors from 16 to 11, thereby decreasing the number of nodes and weights and saving the time and resources required for neural network training and application. The predictability and efficiency were found to greatly improve compared with those previously determined.
This study aimed to classify and predict the risk of diabetes in relation to nutritional intake using a neural network model combined with a sensitivity analysis technique. It revealed that intake data alone could be useful in predicting diabetes risk. Neural network systems may present fluctuations in accuracy due to potential mechanisms such as physical activity, smoking, and genetic characteristics in addition to the nutritional intake applied in this study. Therefore, future studies should integrate risk factors that include the foregoing characteristics.
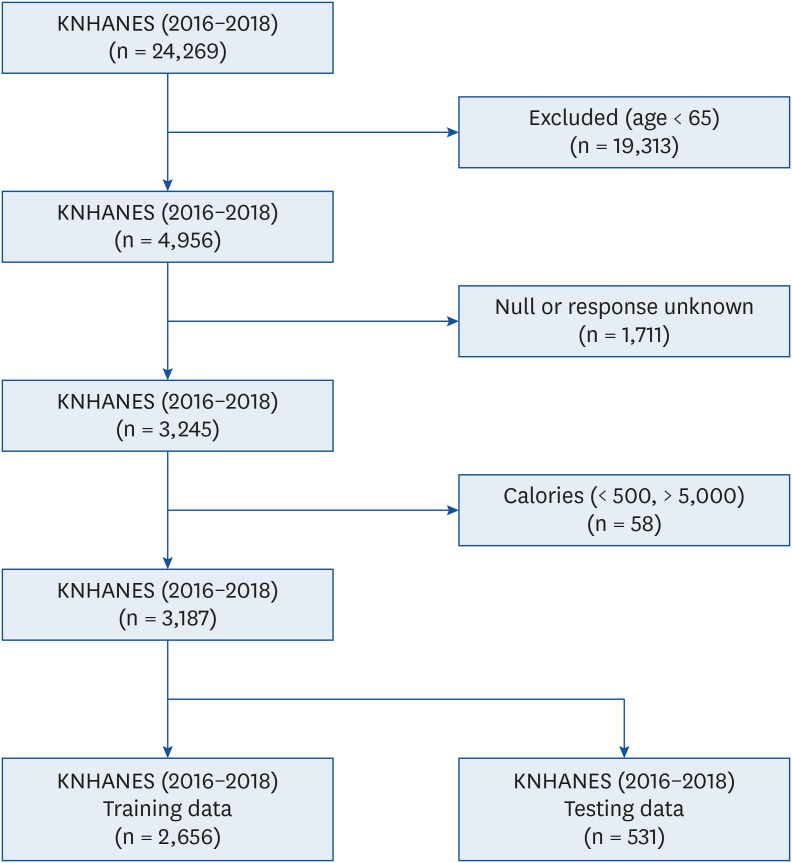
This study is based only on three years of KNHANES data and additional survey results are needed to develop a better performance neural network model for Korean older adults. Further studies utilizing multiple data of the same patient over time could lead to the development of a better neural network model for predicting disease process.




 PDF
PDF Citation
Citation Print
Print



 : The nutrient with the lowest pi at each step, and the corresponding input would be removed in the subsequent step.
: The nutrient with the lowest pi at each step, and the corresponding input would be removed in the subsequent step.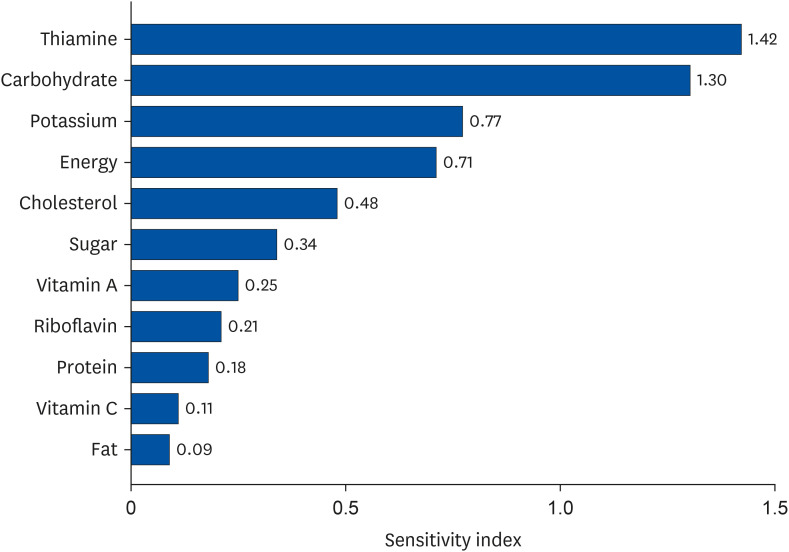
 XML Download
XML Download