Abstract
Purpose
Materials and Methods
Results
Electronic Supplementary Material
Notes
Ethical Statement
The protocol of the present study was approved by the institutional review board of the Severance Hospital, Yonsei University College of Medicine (Seoul, Republic of Korea) (approval no. 4-2012-0900). Informed consent was waived by the institutional review board due to the retrospective study design.
Author Contributions
Conceived and designed the analysis: Kang J, Choi YJ, Kim IK, Lee KY.
Collected the data: Kang J, Choi YJ, Kim H, Baik SH, Kim NK, Lee KY.
Contributed data or analysis tools: Kang J, Choi YJ, Kim IK, Lee HS, Lee KY.
Performed the analysis: Kang J, Lee KY.
Wrote the paper: Kang J, Choi YJ, Lee KY.
Acknowledgments
References
Fig. 1
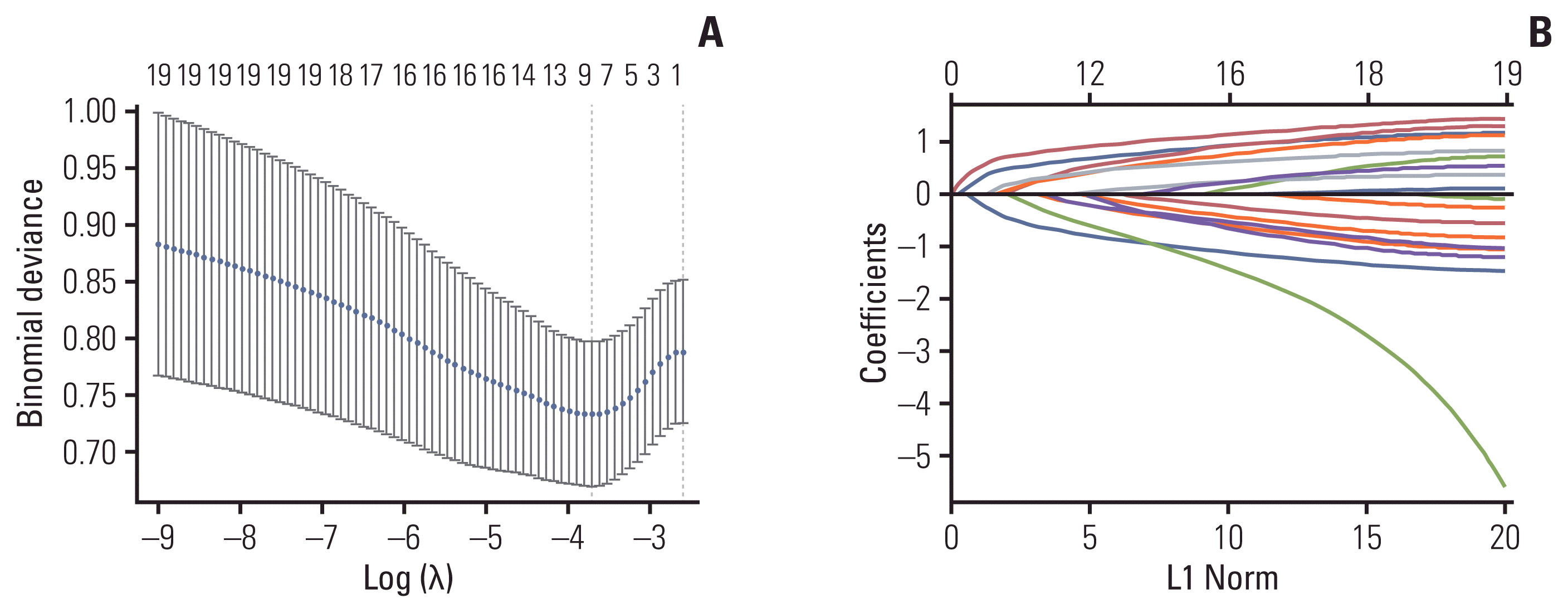
Fig. 2
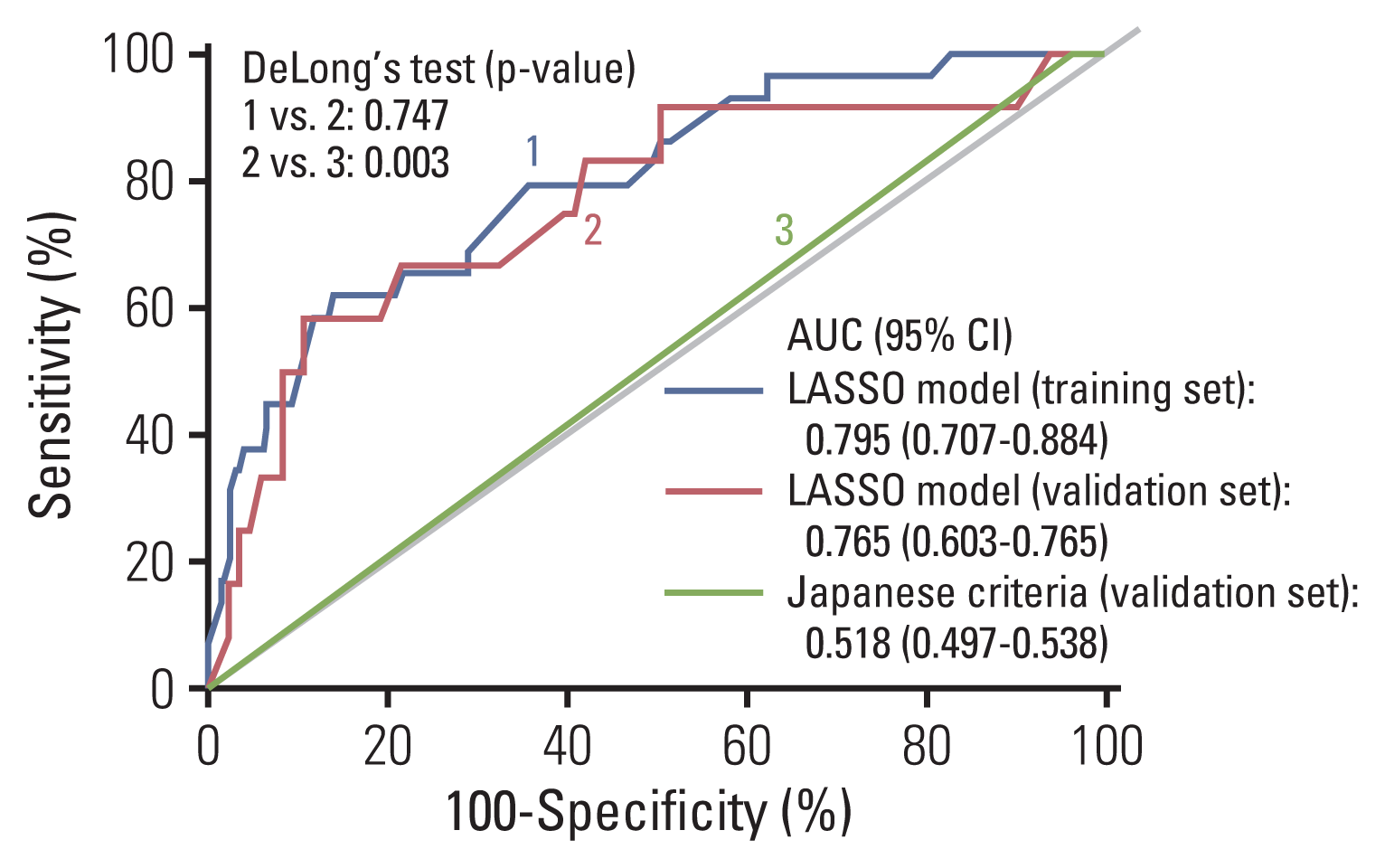
Fig. 3
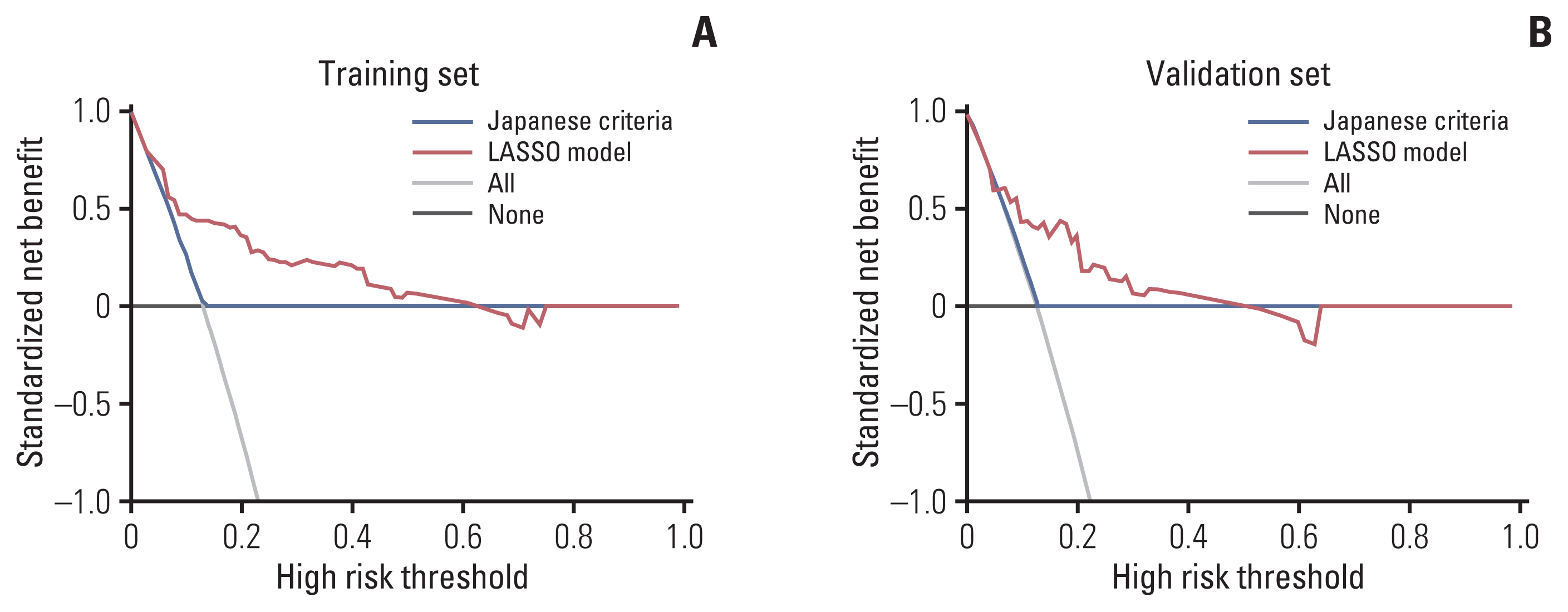
Table 1
| Training set (n=221) | Validation set (n=95) | p-value | |
|---|---|---|---|
| Sex | |||
| Male | 123 (55.7) | 59 (62.1) | 0.347 |
| Female | 98 (44.3) | 36 (37.9) | |
| Age (yr) | |||
| < 70 | 177 (80.1) | 77 (81.1) | 0.966 |
| ≥ 70 | 44 (19.9) | 18 (18.9) | |
| Preoperative CEA (ng/mL) | |||
| < 5.0 | 208 (94.1) | 73 (76.8) | < 0.001 |
| ≥ 5.0 | 13 (5.9) | 22 (23.2) | |
| Tumor location | |||
| Colon | 129 (58.4) | 60 (63.2) | 0.502 |
| Rectum | 92 (41.6) | 35 (36.8) | |
| Retrieved lymph node numbers | 12 (7–17) | 14 (12–21) | < 0.001 |
| Depth of invasion (μm) | |||
| < 1,000 | 20 (9.0) | 6 (6.3) | 0.557 |
| ≥ 1,000 | 201 (91.0) | 89 (93.7) | |
| Histologic gradea) | |||
| G1 | 93 (42.1) | 28 (29.5) | 0.047 |
| G2, G3, etc. | 128 (57.9) | 67 (70.5) | |
| LVI | |||
| Present | 27 (12.2) | 14 (14.7) | 0.668 |
| Absent | 194 (87.8) | 81 (85.3) | |
| Gross morphology | |||
| Pedunculated | 17 (7.7) | 16 (16.8) | 0.025 |
| Flat and sessile | 204 (92.3) | 79 (83.2) | |
| Tumor buddingb) | |||
| Low grade | 88 (39.8) | 38 (40.0) | > 0.99 |
| High grade | 133 (60.2) | 57 (60.0) | |
| Background adenoma | |||
| Present | 196 (88.7) | 84 (88.4) | > 0.99 |
| Absent | 25 (11.3) | 11 (11.6) | |
| LNM | |||
| Negative | 192 (86.9) | 83 (87.4) | > 0.99 |
| Positive | 29 (13.1) | 12 (12.6) | |
| MMR | |||
| MMR-deficient | 14 (6.3) | 8 (8.4) | 0.669 |
| MMR-proficient | 207 (93.7) | 87 (91.6) | |
Table 2
| LN positive, n (%) | Univariable analysis | ||
|---|---|---|---|
| OR (95% CI) | p-value | ||
| Sex | |||
| Female | 14/98 (14.3) | Reference | |
| Male | 15/123 (12.2) | 0.833 (0.37–1.83) | 0.647 |
| Age (yr) | |||
| < 70 | 24/177 (13.6) | Reference | |
| ≥ 70 | 5/33 (11.4) | 0.817 (0.26–2.12) | 0.699 |
| Preoperative CEA (ng/mL) | |||
| < 5.0 | 27/208 (13.0) | Reference | |
| ≥ 5.0 | 2/13 (15.4) | 1.218 (0.18–4.86) | 0.803 |
| Tumor location | |||
| Colon | 14/129 (10.9) | Reference | |
| Rectum | 15/92 (16.3) | 1.6 (0.72–3.53) | 0.239 |
| Depth of invasion (μm) | |||
| < 1,000 | 3/20 (15.0) | Reference | |
| ≥ 1,000 | 26/201 (12.9) | 0.841 (0.25–3.78) | 0.794 |
| Histologic gradea) | |||
| G1 | 7/93 (7.5) | Reference | |
| G2, G3, etc. | 22/128 (17.2) | 2.549 (1.08–6.70) | 0.040 |
| LVI | |||
| Absent | 20/194 (10.3) | Reference | |
| Present | 9/27 (33.3) | 4.35 (1.67–10.84) | 0.001 |
| Gross morphology | |||
| Pedunculated | 2/17 (11.8) | Reference | |
| Flat and sessile | 27/204 (13.2) | 1.144 (0.29–7.51) | 0.863 |
| Tumor buddingb) | |||
| Low grade | 10/133 (7.5) | Reference | |
| High grade | 19/88 (21.6) | 3.387 (1.52–7.97) | 0.003 |
| Background adenoma | |||
| Present | 22/196 (11.2) | Reference | |
| Absent | 7/25 (20.0) | 3.075 (1.09–7.96) | 0.024 |
| MMR | |||
| MMR-deficient | 1/14 (7.1) | Reference | |
| MMR-proficient | 28/207 (13.5) | 2.033 (0.38–37.65) | 0.502 |
| CD3_IM | |||
| Low (< 11.3) | 10/55 (18.2) | Reference | |
| High (≥ 11.3) | 19/166 (11.4) | 0.581 (0.25–1.38) | 0.203 |
| CD3_TC | |||
| Low (< 9.2) | 2/39 (5.1) | Reference | |
| High (≥ 9.2) | 27/182 (14.8) | 3.222 (0.90–20.53) | 0.121 |
| CD8_IM | |||
| Low (< 28.2) | 29/199 (14.6) | Reference | |
| High (≥ 28.2) | 0/27 (0) | 0 (NA-1.68e+26) | 0.990 |
| CD8_TC | |||
| Low (< 18.5) | 24/133 (18.0) | Reference | |
| High (≥ 18.5) | 5/88 (5.7) | 0.273 (0.08–0.69) | 0.011 |
| FOXP3_IM | |||
| Low (< 15.1) | 16/99 (16.2) | Reference | |
| High (≥ 15.1) | 13/122 (10.7) | 0.618 (0.27–1.35) | 0.230 |
| FOXP3_TC | |||
| Low (< 20.3) | 23/157 (14.6) | Reference | |
| High (≥ 20.3) | 6/64 (9.4) | 0.602 (0.21–1.47) | 0.290 |
CEA, carcinoembryonic antigen; CI, confidence interval; IM, invasive margin; LN, lymph node; LVI, lymphovascular invasion; MMR, mismatch repair; NA, not available; OR, odds ratio; TC, tumor center.
Table 3
| Training set (n=221) | Validation set (n=95) | |||
|---|---|---|---|---|
|
|
|
|||
| Japanese criteria vs. LASSO model | p-value | Japanese criteria vs. LASSO model | p-value | |
| NRI (95% CI)a) | 0.722 (0.402–1.041) | < 0.001 | 0.447 (0.041–0.854) | 0.039 |
|
|
||||
| IDI (95% CI) | 0.187 (0.100–0.274) | < 0.001 | 0.121 (0.008–0.234) | 0.034 |




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download