Abstract
Measurable residual disease (MRD) testing, a standard procedure in B-lymphoblastic leukemia (B-ALL) diagnostics, is assessed using multiparametric flow cytometry (MFC) and next-generation sequencing (NGS) analysis of immunoglobulin gene rearrangements. We evaluated the concordance between eight-color, two-tube MFC-MRD the LymphoTrack NGS-MRD assays using 139 follow-up samples from 54 pediatric patients with B-ALL. We also assessed the effect of hemodilution in MFC-MRD assays. The MRD-concordance rate was 79.9% (N=111), with 25 (18.0%) and 3 (2.2%) samples testing positive only by NGS-MRD (MFC−NGS+MRD) and MFC-MRD (MFC+NGS−MRD), respectively. We found a significant correlation in MRD values from total nucleated cells between the two methods (r=0.736 [0.647–0.806], P<0.001). The median MRD value of MFC−NGS+MRD samples was estimated to be 0.0012% (0.0001%–0.0263%) using the NGS-MRD assays. Notably, 14.3% of MFC−NGS+MRD samples showed NGS-MRD values below the limit of detection in the MFC-MRD assays. The percentages of hematogones detected in MFC-MRD assays significantly differed between the discordant and concordant cases (P<0.001). MFC and NGS-MRD assays showed relatively high concordance and correlation in MRD assessment, whereas the NGS-MRD assay detected MRD more frequently than the MFC-MRD assay in pediatric B-ALL. Evaluating the hematogone percentages can aid in assessing the impact of sample hemodilution.
Measurable residual disease (MRD) assessment is a standard practice in B-lymphoblastic leukemia (B-ALL) [1] and serves as a prognostic marker [2]. Multiparametric flow cytometry (MFC)-MRD analysis enables a rapid turnaround time with wide applicability [3, 4]. The sensitivity of MFC-MRD analysis was improved to 10−5 using a two-tube, eight-color next-generation flow MRD, as proposed by the EuroFlow Consortium [5]. Next-generation sequencing (NGS) showed higher sensitivity than MFC-MRD in assaying immunoglobulin gene clonality, was highly concordant with quantitative PCR in detecting immunoglobulin gene rearrangements, and was less laborious while providing comprehensive results [6]. Various MFC-MRD methods, involving different antibody–fluorophore combinations, sample-processing steps, and data analysis, have been developed; however, relatively few NGS-MRD assays, such as the LymphoTrack (Invivoscribe, San Diego, CA, USA) [7] and clonoSEQ (Adaptive Biotechnologies, Washington, USA) assays, are available for detecting immunoglobulin gene rearrangements [8]. MFC-MRD and NGS-MRD assays are complementary, and patients who are MRD-negative by both methods have better treatment outcomes [6, 9]. However, few studies have compared the MFC-MRD and NGS-MRD assays [6, 10]. Therefore, we compared MFC-MRD and NGS-MRD results in pediatric patients with B-ALL and evaluated the impact of hemodilution, especially in peripheral blood samples.
We examined bone marrow (BM) follow-up samples from pediatric patients with B-ALL via simultaneous MFC-MRD and LymphoTrack immunoglobulin heavy chain gene (IGH) analysis (Invivoscribe), using samples obtained in Seoul National University Hospital and Seoul National University Bundang Hospital from February 2022 to December 2022. The eight-color, two-tube MFC-MRD panel was modified based on EuroFlow B-ALL MRD guidelines [4], with common markers used in both tubes (CD34-PerCPCy5.5, CD19-PECy-7, CD10-APC, CD20-V450, CD38-APC-H7, and CD45-V500c). CD81-FITC and C73-PE were included in tube one only, and CD58-FITC, CD66c, and CD123-PE were included in tube two only. The antibodies were obtained from Becton Dickson (BD) and Company (San Diego, CA, USA), and validation results are presented in the Supplemental Materials. For patients treated with anti-CD19 therapy, CD22-FITC, CD66b-PE, and CD24-PECy7 were added to the second tube [11]. We conducted bulk lysis following the EuroFlow guidelines, acquiring 5 million events for each tube or as many events as possible (when minimal cells were available), using a BD FACSCanto (BD) instrument and Infinicyt Software 2.0.5 (Cytognos, Salamanca, Spain). The limit of detection (LOD) was defined as 60 events/100 total nucleated cells (TNCs), with a lower limit of quantitation (LLOQ) as 100 events/100 TNCs. Hemodilution was assessed based on quantitating nucleated red blood cells (RBCs) and identifying hematogones [12]. To identify nucleated RBCs, we identified cells that lacked all included markers and exhibited low forward and side scatter. Hematogones were identified as CD19+, CD38+, CD45dim, CD34+/−, and CD10+ cells, and their percentages were determined based on the number of TNCs in the sample. This study was approved by our local ethics committee (approval number B-2301-805-104).
For NGS-MRD, we extracted DNA from BM aspirates with a QIAsymphony DSP DNA Midi Kit using the QIAsymphony system (Qiagen, Hilden, Germany). IGH rearrangement and clones were assessed using the LymphoTrack MRD-NGS assay (Invivoscribe). We tested 24 samples in a single run with 800 ng of DNA, achieving a minimum sensitivity of 10−4. The data were processed using LymphoTrack Analysis Software and the MRD Data Analysis Tool (Invivoscribe). NGS-MRD provided clonal/TNC (%) for MRD among TNCs and clonal/total IGH read count (%) for MRD among B cells. The TNC-based percentage was calculated using the LymphoQuant B-cell internal control (Invivoscribe).
We compared the patients’ clinical characteristics using the Mann–Whitney U-test and analyzed the correlation between the MFC-MRD and NGS-MRD results using Spearman’s correlation coefficient. To compare the median positive MRD values, we performed the Wilcoxon matched-pairs signed-rank test. Statistical analyses were performed using GraphPad Prism 9.5.1 (GraphPad Software Inc., San Diego, CA, USA) and R Software version 3.6.1.
We assessed 139 samples from 54 pediatric patients with B-ALL (median age: 9 yrs; range: 1–18 yrs). Table 1 summarizes the patient and sample characteristics. Among these patients, 40.7% (N=22) had B-ALL with recurrent genetic abnormalities, and all patients had clonal immunoglobulin gene rearrangements identified at diagnosis, with a median of two types of clones (range: 1–4). Among the 139 samples, MRD assessment was conducted after anti-CD19 therapy (such as blinatumomab or chimeric antigen receptor T cell therapy). Alternative B-cell markers were used for 18.0% (N=25) samples from seven patients.
Thirty-three (23.7%) samples from 15 patients were positive for MRD using MFC-MRD (median: 0.0180% [0.0007%–19.6%]), and 55 (39.6%) samples from 25 patients were positive for MRD using NGS-MRD (median: 0.0229% [0.0001%–112.8%]). Regarding the MFC-MRD analysis, a median of 6,900,221 (916,235–9,302,173) TNC events were analyzed, with a median LOD of 0.0009% (0.0006%–0.0066%) and a median LLOQ of 0.0014% (0.0002%–0.0109%). Regarding the NGS-MRD analysis, a median total IGH read count of 1,087,179 (1,510–2,625,491) was analyzed.
The concordance rate between both methods was 79.9% (N=111) with the 139 samples analyzed; 30 and 81 samples were MRD-positive and MRD-negative, respectively, based on both methods. Further, 25 (18.0%) and three (2.2%) samples were positive based on NGS-MRD only (MFC−NGS+MRD) and MFC-MRD only (MFC+NGS−MRD), respectively (Fig. 1). A moderate correlation was found between both methods for MRD values from TNCs (r=0.736 [0.647–0.806], P<0.001) and for MRD percentages among B cells (r=0.748 [0.662–0.815], P<0.001). Additionally, a strong correlation was identified between the MRD values based on TNCs and B cells obtained via MFC analysis (r=0.915 [0.881–0.939], P<0.001) or NGS analysis (r=0.991 [0.987–0.994], P<0.001). When considering only positive MRD values, a significant difference was noted in the median values, with MFC-MRD analysis revealing a 0.120% lower median MRD value than NGS-MRD analysis (P<0.001).
Among the 28 samples with discordant MRD values, 25 MFC−NGS+MRD samples (N=20 patients) showed a median MRD value of 0.0035% (0.0001%–0.5250%) by NGS-MRD, with 24.0% of the MFC−NGS+MRD samples showing an NGS-MRD value below the LLOQ of the MFC-MRD assay. Ten MFC−NGS+MRD samples showed negative MFC-MRD results in the interval before the NGS-MRD results were reported as being negative. Regarding the three MFC+NGS−MRD samples, the median MRD value was 0.0026% (0.0025%–0.0089%) by MFC-MRD. Notably, one sample with MFC+NGS−MRD showed a positive BCR::ABL1 quantitative PCR result, consistent with the MFC-MRD findings (Supplemental Data Table S1).
We assessed whether MFC-MRD analysis involving gating for alternative B-cell markers would result in more discrepant cases and found discrepant MRD values for 28.0% (N=7) and 18.4% (N=21) of samples from patients treated with anti-CD19 therapy and conventional therapies, respectively. However, these discrepancies did not significantly differ between the two groups (P=0.281).
A median of 0.0051% (0–4.3300%) of hematogones was present in the MFC-MRD-positive samples. The median percentage of hematogones was 0.0000% (N=28) in discrepant cases and 0.0109% (N=111) in concordant cases (P=0.021). The optimal cutoff value for predicting concordant MRD was 0.0555% for the hematogones, with an area under the curve of 0.637 (P<0.001).
MFC-MRD has a history of use, whereas NGS-MRD is a more recent option for detecting B-ALL. We compared the performance of the MFC-MRD assay with that of a commercial NGS-based LymphoTrack IGH assay for pediatric patients with B-ALL. Concordance between the MFC-MRD and NGS-MRD assays was high and similar to previous findings showing more frequency and sensitive MRD detection with NGS-MRD [8, 9, 13].
Many discordant cases (85.7%, N=24) had MRD values below 0.01%, and the concordance rate was higher (92.8%) when the cutoff was adjusted to 0.01%, a well-validated value [2]. Regarding the MFC−NGS+MRD cases, the MRD values detected using NGS-MRD were below the LOD of the MFC-MRD assay in 14.3% (N=4) of the cases, rendering them undetectable by MFC-MRD. Most MFC−NGS+MRD cases were detected after the MFC-MRD results had turned negative, often within one measurement interval, indicative of a more sensitive MRD detection capability. Although several MFC−NGS+MRD cases were within the detection range of the MFC-MRD assay, immunophenotypic changes may occur, and the emergence of leukemic cells with a more mature immunophenotype following therapy may hinder detection by MFC-MRD [8, 14].
The MRD values were higher with the NGS-MRD assay, possibly because of normalization with the number of TNCs as an internal control [15], whereas MFC-MRD analysis might have inadvertently included cellular debris as nucleated cell events, as some methods incorporate nucleated acid dye [16]. MRD values based on TNC or B-cell normalization correlated significantly, and the clinical significance of the normalized values obtained with the LymphoTrack NGS-MRD [15] and clonoSEQ [8] has been reported, although further validation might be required.
MRD results are significantly influenced by the sample quality and pre-analytic conditions, with more notable effects observed on MFC-MRD results than on NGS-MRD results [17]. We assessed hemodilution via MFC-MRD analysis [12], where the percentage of hematogones, rather than the percentage of nucleated RBCs, was used to assess hemodilution. In the future, combinations of different BM cell types may be used to assess hemodilution [18].
However, as with many studies regarding MFC-MRD and NGS-MRD analysis of B-ALL [8, 13], some patients lack identifiable clones with NGS at diagnosis [19]; thus, NGS-MRD may not be feasible, and IGH-clonality results may not be available for referred patients, making MFC-MRD more reliable.
A major limitation of this study is the relatively short follow-up period (median: 14.3 months; range: 5.1–88.5 months). Survival analysis could not be performed, rendering it difficult to determine the false-positive and false-negative rates of both assays. Future studies should be conducted to assess the significance of low-level MRD positivity. Although some studies have compared NGS-MRD analysis with clonoSEQ and MFC-MRD analysis [7, 13], relatively few studies have compared LymphoTrack NGS-MRD and MFC-MRD assays using samples from patients with B-ALL [15].
In conclusion, the MFC-MRD and NGS-MRD results correlated well when the MRD values were based on either B cells or TNCs. NGS-MRD analysis showed a higher rate of MRD detection than MFC-MRD using samples from pediatric patients with B-ALL. Further, evaluating the percentages of hematogones can help in assessing the effects of hemodilution during MFC-MRD analysis.
Supplementary materials can be found via https://doi.org/10.3343/alm.2023.0412
Notes
AUTHOR CONTRIBUTIONS
Hwang SM conceived the study, designed the experiment, analyzed the data, and wrote the manuscript. Oh I, Kwon SR, Lee JS, and Seong MW analyzed the data, and Lee JS analyzed the data and wrote the manuscript. All authors contributed to the manuscript and approved the submitted version.
References
1. Brown P, Inaba H, Annesley C, Beck J, Colace S, Dallas M, et al. 2020; Pediatric acute lymphoblastic leukemia, version 2.2020, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 18:81–112. DOI: 10.6004/jnccn.2020.0001. PMID: 31910389.

2. Berry DA, Zhou S, Higley H, Mukundan L, Fu S, Reaman GH, et al. 2017; Association of minimal residual disease with clinical outcome in pediatric and adult acute lymphoblastic leukemia: a meta-analysis. JAMA Oncol. 3:e170580. DOI: 10.1001/jamaoncol.2017.0580. PMID: 28494052. PMCID: PMC5824235.
3. Cherian S, Soma LA. 2021; How I diagnose minimal/measurable residual disease in B lymphoblastic leukemia/lymphoma by flow cytometry. Am J Clin Pathol. 155:38–54. DOI: 10.1093/ajcp/aqaa242. PMID: 33236071.

4. Kim HY, Yoo IY, Lim DJ, Kim HJ, Kim SH, Yoon SE, et al. 2022; Clinical utility of next-generation flow-based minimal residual disease assessment in patients with multiple myeloma. Ann Lab Med. 42:558–65. DOI: 10.3343/alm.2022.42.5.558. PMID: 35470273. PMCID: PMC9057816.

5. Theunissen P, Mejstrikova E, Sedek L, van der Sluijs-Gelling AJ, Gaipa G, Bartels M, et al. 2017; Standardized flow cytometry for highly sensitive MRD measurements in B-cell acute lymphoblastic leukemia. Blood. 129:347–57. DOI: 10.1182/blood-2016-07-726307. PMID: 27903527. PMCID: PMC5291958.

6. Wood B, Wu D, Crossley B, Dai Y, Williamson D, Gawad C, et al. 2018; Measurable residual disease detection by high-throughput sequencing improves risk stratification for pediatric B-ALL. Blood. 131:1350–9. DOI: 10.1182/blood-2017-09-806521. PMID: 29284596. PMCID: PMC5865233.

7. Paulsen K, Marincevic M, Cavelier L, Hollander P, Amini RM. 2022; LymphoTrack is equally sensitive as PCR GeneScan and Sanger sequencing for detection of clonal rearrangements in ALL patients. Diagnostics (Basel). 12:1389. DOI: 10.3390/diagnostics12061389. PMID: 35741199. PMCID: PMC9222020. PMID: 59daed8511b247a0a966a0b0a83c5d19.

8. Wu D, Emerson RO, Sherwood A, Loh ML, Angiolillo A, Howie B, et al. 2014; Detection of minimal residual disease in B lymphoblastic leukemia by high-throughput sequencing of IGH. Clin Cancer Res. 20:4540–8. DOI: 10.1158/1078-0432.CCR-13-3231. PMID: 24970842. PMCID: PMC5142743.
9. Short NJ, Kantarjian H, Ravandi F, Konopleva M, Jain N, Kanagal-Shamanna R, et al. 2022; High-sensitivity next-generation sequencing MRD assessment in ALL identifies patients at very low risk of relapse. Blood Adv. 6:4006–14. DOI: 10.1182/bloodadvances.2022007378. PMID: 35533262. PMCID: PMC9278301.

10. Cheng S, Inghirami G, Cheng S, Tam W. 2018; Simple deep sequencing-based post-remission MRD surveillance predicts clinical relapse in B-ALL. J Hematol Oncol. 11:105. DOI: 10.1186/s13045-018-0652-y. PMID: 30134947. PMCID: PMC6103872. PMID: d7da29e8787e4610b0cda54e76957a87.

11. Cherian S, Miller V, McCullouch V, Dougherty K, Fromm JR, Wood BL. 2018; A novel flow cytometric assay for detection of residual disease in patients with B-lymphoblastic leukemia/lymphoma post anti-CD19 therapy. Cytometry B Clin Cytom. 94:112–20. DOI: 10.1002/cyto.b.21482. PMID: 27598971.

12. Flores-Montero J, Sanoja-Flores L, Paiva B, Puig N, García-Sánchez O, Böttcher S, et al. 2017; Next generation flow for highly sensitive and standardized detection of minimal residual disease in multiple myeloma. Leukemia. 31:2094–103. DOI: 10.1038/leu.2017.29. PMID: 28104919. PMCID: PMC5629369.

13. Momen N, Tario J, Fu K, Qian YW. 2023; Multiparameter flow cytometry and ClonoSEQ correlation to evaluate precursor B-lymphoblastic leukemia measurable residual disease. J Hematopathol. 16:85–94. DOI: 10.1007/s12308-023-00544-9. PMID: 38175444.

14. Chen W, Karandikar NJ, McKenna RW, Kroft SH. 2007; Stability of leukemia-associated immunophenotypes in precursor B-lymphoblastic leukemia/lymphoma: a single institution experience. Am J Clin Pathol. 127:39–46. DOI: 10.1309/7R6MU7R9YWJBY5V4. PMID: 17145625.

15. Lee JW, Kim Y, Ahn A, Lee JM, Yoo JW, Kim S, et al. 2022; Clinical implication of minimal residual disease assessment by next-generation sequencing-based immunoglobulin clonality assay in pediatric B-acute lymphoblastic leukemia. Front Oncol. 12:957743. DOI: 10.3389/fonc.2022.957743. PMID: 36185293. PMCID: PMC9521036. PMID: 980933830448449e973d16bb75c2529b.

16. Dworzak MN, Gaipa G, Ratei R, Veltroni M, Schumich A, Maglia O, et al. 2008; Standardization of flow cytometric minimal residual disease evaluation in acute lymphoblastic leukemia: multicentric assessment is feasible. Cytometry B Clin Cytom. 74:331–40. DOI: 10.1002/cyto.b.20430. PMID: 18548617.

17. Kriegsmann K, Hundemer M, Hofmeister-Mielke N, Reichert P, Manta CP, Awwad MHS, et al. 2020; Comparison of NGS and MFC methods: Key metrics in multiple myeloma MRD assessment. Cancers (Basel). 12:2322. DOI: 10.3390/cancers12082322. PMID: 32824635. PMCID: PMC7464347. PMID: 11561f474ce44996ac5ae432665b4577.

18. Hoffmann J, Thrun MC, Röhnert MA, von Bonin M, Oelschlägel U, Neubauer A, et al. 2023; Identification of critical hemodilution by artificial intelligence in bone marrow assessed for minimal residual disease analysis in acute myeloid leukemia: the Cinderella method. Cytometry A. 103:304–12. DOI: 10.1002/cyto.a.24686. PMID: 36030398.

19. Mai H, Li Q, Wang G, Wang Y, Liu S, Tang X, et al. 2023; Clinical application of next-generation sequencing-based monitoring of minimal residual disease in childhood acute lymphoblastic leukemia. J Cancer Res Clin Oncol. 149:3259–66. DOI: 10.1007/s00432-022-04151-6. PMID: 35918464.

Fig. 1
Comparison of MRD data obtained using MFC-MRD and NGS-MRD. The grid in the lower right of the graph shows the numbers of NGS+/MFC−, NGS+/MFC+, NGS−/MFC+, and NGS−/MFC−MRD samples.
Abbreviations: MRD, measurable residual disease; MFC, multiparametric flow cytometry; NGS, next-generation sequencing; TNC, total nucleated cells.
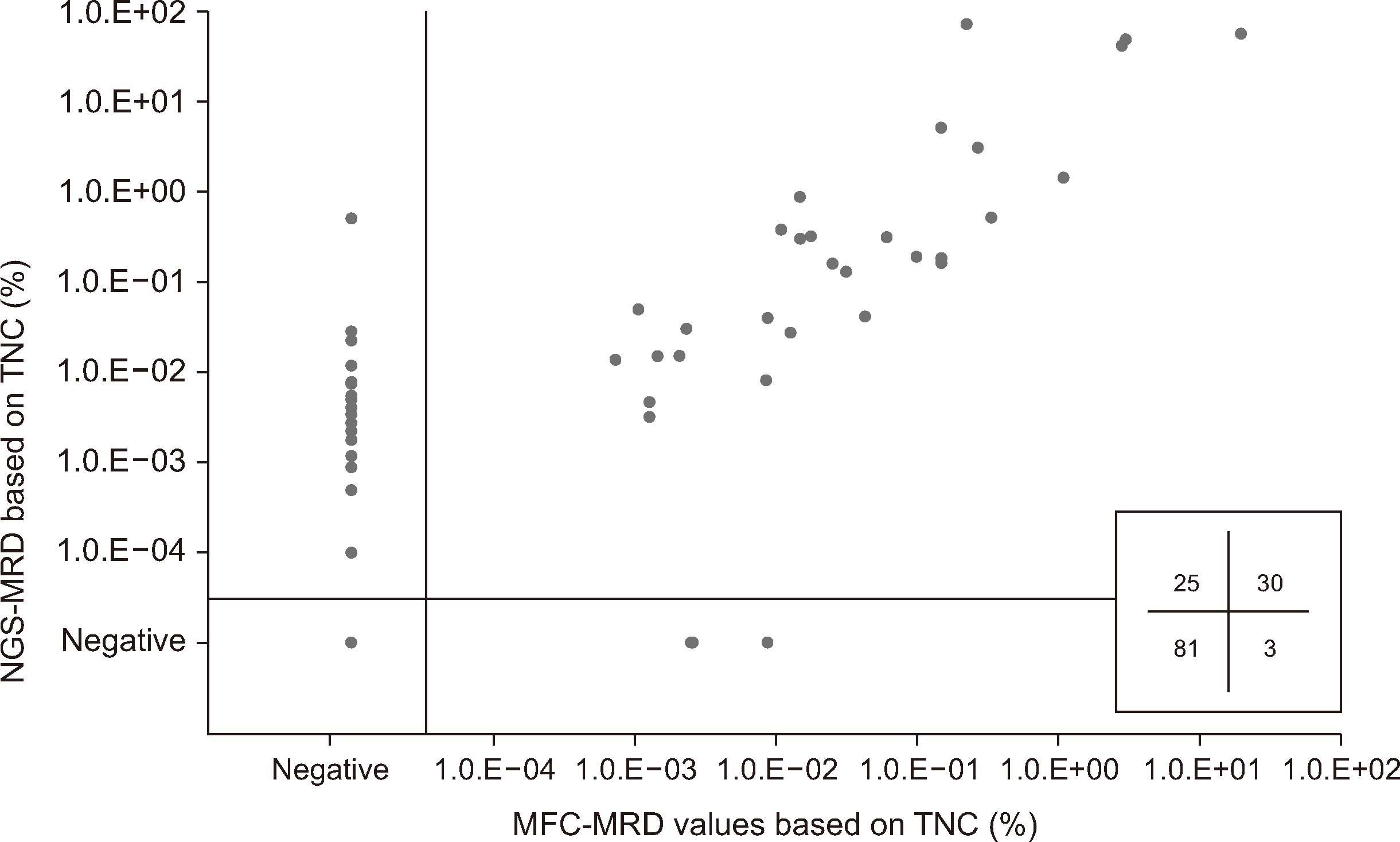
Table 1
Patient and sample characteristics




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download