1. Nyberg DA, Souter VL. Sonographic markers of fetal trisomies: second trimester. J Ultrasound Med. 2001; 20(6):655–674. PMID:
11400940.
2. Society for Maternal-Fetal Medicine (SMFM). Electronic address: pubs@smfm.org. Prabhu M, Kuller JA, Biggio JR. Society for Maternal-Fetal Medicine consult series #57: evaluation and management of isolated soft ultrasound markers for aneuploidy in the second trimester: (replaces consults #10, single umbilical artery, October 2010; #16, isolated echogenic bowel diagnosed on second-trimester ultrasound, August 2011; #17, evaluation and management of isolated renal pelviectasis on second-trimester ultrasound, December 2011; #25, isolated fetal choroid plexus cysts, April 2013; #27, isolated echogenic intracardiac focus, August 2013). Am J Obstet Gynecol. 2021; 225(4):B2–15.
3. Cunningham FG, Leveno KJ, Dashe JS, Hoffman BL, Spong CY, Casey BM. Williams Obstetrics. 26th ed. New York, NY, USA: McGraw-Hill Medical;2022.
4. Ko HS, Kwak DW, Oh SY, Choi SK, Hong JS, Hwang HS, et al. Clinical significance of soft markers in second trimester ultrasonography for pregnant Korean women: a multicenter study and literature review. Obstet Gynecol Sci. 2022; 65(2):145–155. PMID:
35184524.
5. Bromley B, Lieberman E, Shipp TD, Benacerraf BR. The genetic sonogram: a method of risk assessment for down syndrome in the second trimester. J Ultrasound Med. 2002; 21(10):1087–1096. PMID:
12369663.
6. American College of Obstetricians and Gynecologists’ Committee on Practice Bulletins—Obstetrics. Committee on Genetics. Society for Maternal-Fetal Medicine. Screening for fetal chromosomal abnormalities: ACOG practice bulletin, number 226. Obstet Gynecol. 2020; 136(4):e48–e69. PMID:
32804883.
7. Lichtenbelt KD, Knoers NV, Schuring-Blom GH. From karyotyping to array-CGH in prenatal diagnosis. Cytogenet Genome Res. 2011; 135(3-4):241–250. PMID:
22086062.
8. Manning M, Hudgins L. Professional Practice and Guidelines Committee. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med. 2010; 12(11):742–745. PMID:
20962661.
9. Michelson DJ, Shevell MI, Sherr EH, Moeschler JB, Gropman AL, Ashwal S. Evidence report: genetic and metabolic testing on children with global developmental delay: report of the quality standards subcommittee of the American Academy of Neurology and the practice committee of the Child Neurology Society. Neurology. 2011; 77(17):1629–1635. PMID:
21956720.
10. American College of Obstetricians and Gynecologists Committee on Genetics. Committee opinion No. 581: the use of chromosomal microarray analysis in prenatal diagnosis. Obstet Gynecol. 2013; 122(6):1374–1377. PMID:
24264715.
11. Norton ME. Callen's Ultrasonography in Obstetrics and Gynecology E-Book: Callen's Ultrasonography in Obstetrics and Gynecology E-Book. Amsterdam, The Netherlands: Elsevier Health Sciences;2016.
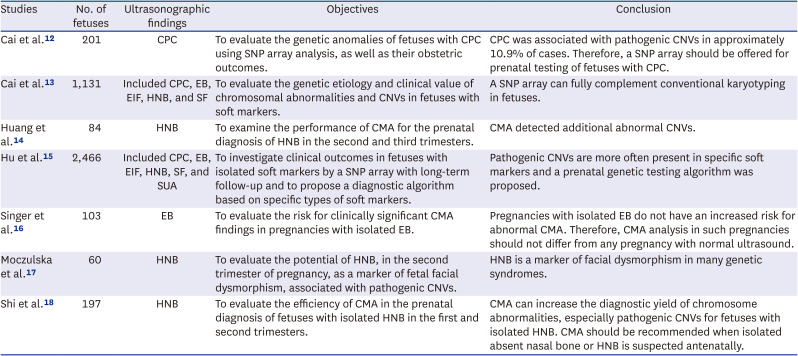
12. Cai M, Huang H, Su L, Wu X, Xie X, Xu L, et al. Choroid plexus cysts: single nucleotide polymorphism array analysis of associated genetic anomalies and resulting obstetrical outcomes. Risk Manag Healthc Policy. 2021; 14:2491–2497. PMID:
34163268.
13. Cai M, Lin N, Chen X, Fu M, Guo N, Xu L, et al. Evaluation of chromosomal abnormalities and copy number variations in fetuses with ultrasonic soft markers. BMC Med Genomics. 2021; 14:19. PMID:
33435955.
14. Huang H, Cai M, Ma W, Lin N, Xu L. Chromosomal microarray analysis for the prenatal diagnosis in fetuses with nasal bone hypoplasia: a retrospective cohort study. Risk Manag Healthc Policy. 2021; 14:1533–1540. PMID:
33889037.
15. Hu T, Tian T, Zhang Z, Wang J, Hu R, Xiao L, et al. Prenatal chromosomal microarray analysis in 2466 fetuses with ultrasonographic soft markers: a prospective cohort study. Am J Obstet Gynecol. 2021; 224(5):516.e1–516.16.
16. Singer A, Maya I, Koifman A, Nasser Samra N, Baris HN, Falik-Zaccai T, et al. Microarray analysis in pregnancies with isolated echogenic bowel. Early Hum Dev. 2018; 119:25–28. PMID:
29522884.
17. Moczulska H, Serafin M, Wojda K, Borowiec M, Sieroszewski P. Fetal nasal bone hypoplasia in the second trimester as a marker of multiple genetic syndromes. J Clin Med. 2022; 11(6):1513. PMID:
35329839.
18. Shi X, Lu J, Li L, Wei R, Wu J. Prenatal chromosomal microarray analysis in foetuses with isolated absent or hypoplastic nasal bone. Ann Med. 2022; 54(1):1297–1302. PMID:
35506821.
19. Chang H, Li L, Li M, Xiao X. Rare and common variants at 16p11.2 are associated with schizophrenia. Schizophr Res. 2017; 184:105–108. PMID:
27889382.
20. D’Angelo D, Lebon S, Chen Q, Martin-Brevet S, Snyder LG, Hippolyte L, et al. Defining the effect of the 16p11.2 duplication on cognition, behavior, and medical comorbidities. JAMA Psychiatry. 2016; 73(1):20–30. PMID:
26629640.
21. Maillard AM, Ruef A, Pizzagalli F, Migliavacca E, Hippolyte L, Adaszewski S, et al. The 16p11.2 locus modulates brain structures common to autism, schizophrenia and obesity. Mol Psychiatry. 2015; 20(1):140–147. PMID:
25421402.
22. Steinman KJ, Spence SJ, Ramocki MB, Proud MB, Kessler SK, Marco EJ, et al. 16p11.2 deletion and duplication: characterizing neurologic phenotypes in a large clinically ascertained cohort. Am J Med Genet A. 2016; 170(11):2943–2955. PMID:
27410714.
23. Maya I, Sharony R, Yacobson S, Kahana S, Yeshaya J, Tenne T, et al. When genotype is not predictive of phenotype: implications for genetic counseling based on 21,594 chromosomal microarray analysis examinations. Genet Med. 2018; 20(1):128–131. PMID:
28726807.
24. Hassfurther A, Komini E, Fischer J, Leipoldt M. Clinical and genetic heterogeneity of the 15q13. 3 microdeletion syndrome. Mol Syndromol. 2016; 6(5):222–228. PMID:
26997942.
25. Pizzo L, Andrieux J, Amor DJ, Girirajan S. Clinical utility gene card for: 16p12.2 microdeletion. Eur J Hum Genet. 2017; 25(2):271.
26. Kirov G, Rees E, Walters JT, Escott-Price V, Georgieva L, Richards AL, et al. The penetrance of copy number variations for schizophrenia and developmental delay. Biol Psychiatry. 2014; 75(5):378–385. PMID:
23992924.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download