Abstract
The TRPM4 gene encodes a Ca2+-activated monovalent cation channel called transient receptor potential melastatin 4 (TRPM4) that is expressed in various tissues. Dysregulation or abnormal expression of TRPM4 has been linked to a range of diseases. We introduced the hemagglutinin (HA) tag into the extracellular S6 loop of TRPM4, resulting in an HA-tagged version called TRPM4-HA. This TRPM4-HA was developed to investigate the purification, localization, and function of TRPM4 in different physiological and pathological conditions. TRPM4-HA was successfully expressed in the intact cell membrane and exhibited similar electrophysiological properties, such as the current-voltage relationship, rapid desensitization, and current size, compared to the wild-type TRPM4. The presence of the TRPM4 inhibitor 9-phenanthrol did not affect these properties. Furthermore, a wound-healing assay showed that TRPM4-HA induced cell proliferation and migration, similar to the native TRPM4. Co-expression of protein tyrosine phosphatase, non-receptor type 6 (PTPN6 or SHP-1) with TRPM4-HA led to the translocation of TRPM4-HA to the cytosol. To investigate the interaction between PTPN6 and tyrosine residues of TRPM4 in enhancing channel activity, we generated four mutants in which tyrosine (Y) residues were substituted with phenylalanine (F) at the N-terminus of TRPM4. The YF mutants displayed properties and functions similar to TRPM4-HA, except for the Y256F mutant, which showed resistance to 9-phenanthrol, suggesting that Y256 may be involved in the binding site for 9-phenanthrol. Overall, the creation of HA-tagged TRPM4 provides researchers with a valuable tool to study the role of TRPM4 in different conditions and its potential interactions with other proteins, such as PTPN6.
Transient receptor potential melastatin 4 (TRPM4) is a Ca2+-activated, nonselective monovalent cationic channel that does not allow the passage of divalent Ca2+ or Mg2+ ions [1,2]. Its expression is observed in various organs and tissues throughout the body [3-6]. TRPM4 plays a crucial role in regulating diverse cellular processes, including cell migration, proliferation, and differentiation [7-9]. Under normal physiological conditions, TRPM4 interacts with focal adhesion molecules involved in cell assembly and adhesion processes, thereby contributing to cell migration [10]. This interaction occurs in dendritic and immune cells, including T lymphocytes and mast cells, and contributes to cellular functions [11-15]. Due to its widespread expression and diverse functional roles in different cell types, TRPM4 has been implicated in the pathophysiology of various cardiovascular and immune diseases [16-18].
In recent years, research on TRPM4 has extended to its potential involvement in different types of cancer. Studies have shown that TRPM4 is overexpressed in several cancer cell lines, including HeLa cells (cervical cancer), as well as cells from prostate, colorectal, liver, breast, urinary bladder, and endometrial cancers [15,19-22]. The overexpression of TRPM4 has been associated with key cancer functions such as increased cell proliferation, migration, and alterations in the cell cycle.
To investigate the diverse functions of TRPM4 and gain a better understanding of its potential contribution to various pathophysiological conditions, it is essential to have a reliable tool or method for accurately detecting its expression. Additionally, understanding its trafficking and degradation can provide valuable insights into the regulation of TRPM4 and its involvement in various cellular processes.
Epitope tagging of TRPM4 is a useful strategy for studying its channel trafficking and expression. While previous studies have inserted an epitope at either the amino (N) or carboxy (C) terminus of TRPM4, this study suggests that the N-terminus is a more promising site for epitope insertion, as attempts to tag the C-terminus have been unsuccessful for other proteins like protein tyrosine phosphatase, non-receptor type 6 (PTPN6) [23]. To address this issue, we designed a mutant by inserting an epitope motif between the fifth (S5) and sixth (S6) transmembrane domains, allowing it to protrude from the cell membrane [24,25]. Epitope tagging has become a common approach for investigating various molecular aspects of proteins, including size, abundance, trafficking patterns, cellular localization, and other applications [26-29].
Considering the ease of insertion and detection, we selected a hemagglutinin (HA) tag, which is a nine-amino acid sequence (YPYDVPDYA) that shares two amino acids located in the extracellular region. This tag can be readily detected using a specific antibody without the need for membrane permeabilization with a detergent [30-32]. This strategy was adopted from previous studies on the heteromeric assembly of TRPC3 with TRPC4 [33]. However, it remains unknown whether the HA tag attached to TRPM4 disrupts its structure, expression, or channel function. Therefore, this study aims to determine whether TRPM4-HA can safely replace the endogenous protein and serve as a reliable marker in TRPM4-related research on pathophysiological conditions and diseases.
The TRPM4 blockers 9-phenanthrol and N-Methyl-D-glucamine (NMDG) were purchased from Sigma–Aldrich.
cDNA encoding human TRPM4 (GenBank accession no. NM_017636) in EGFP-IRES vector was used to generate HA epitope-tagged TRPM4. For HA epitope tagging in the extracellular S6 domain of TRPM4 between amino acids Y1015 and A1016, the nucleotide sequence encoding HA (YPYDVPDYA) was incorporated into the primers by site-directed mutagenesis (EZchange Site-directed Mutagenesis kit; Enzynomics). The primers were designed based on the reported sequence of human TRPM4; Forward primer 5’-TTCCAGATTATGCCAACTGGCTGGTGGTGCTGCT-3’ and Reverse primer 5’-CATCGTATGGAGTACTGGGAGACGCAGGTGCCCG-3.’
The cytosolic PTPN6, interacts with and dephosphorylates TRPM4, thereby increasing the activity of TRPM4 [23]. To confirm whether PTPN6 is required for activation of TRPM4, pSicoR-PTPN6-shRNA was generated using oligonucleotide-directed mutagenesis (Enzynomics). Nucleotides 1148–1169 (5’-GGAGCATGACACAACCGAATA-3’) in human PTPN6 (GenBank accession no. NM_002831) were selected as a target shRNA. Human PTPN6-shRNA was validated by confirming the sequence and then cloned into adenoviral vectors using the Gateway cloning system (Thermo Fisher Scientific).
To confirm the interaction between PTPN6 and the tyrosine residues of TRPM4, phosphorylation-incompetent TRPM4-HA mutants were generated. Tyrosine phosphorylation sites were predicted using NetPhos-3.1 (https://services.healthtech.dtu.dk/service.php?NetPhos-3.1). Four Tyr (Y) residues in the N-terminal region of TRPM4 (residues 75, 256, 393, and 519) were identified as putative phosphorylation sites. These four residues have been predicted as potential binding sites for PTPN6. Site-directed mutagenesis (Enzynomics) was performed to change the Tyr (Y) at residues 75, 256, 393, or 519 to Phe (F), which are referred to as YF mutants in the current study.
The HEK293 cell line was purchased from the Korean Cell Line Bank, and the cells were maintained in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% FBS and 1% penicillin-streptomycin in an incubator at 37°C in 95% O₂ and 5% CO₂. The cells were transiently transfected with cDNAs using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific) according to the manufacturer’s recommended protocol.
HEK293 cells were transfected with TRPM4-HA or TRPM4-HA-YF mutants in the absence or presence of PTPN6-shRNA. After 48 h of incubation, the cells were harvested and lysed using a Membrane Protein Extraction Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The subcellular fractions were obtained by separating the cell membrane from the cytoplasm. The concentrations of solubilized proteins in the extracts were determined by BCA Protein Assay (Thermo Fisher Scientific). Proteins in the extracts (10 µg) were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The separated proteins were then transferred onto methanol-activated polyvinylidene difluoride membranes (Merck). The membranes were blocked with 5% skim milk in 1× TBST and washed three times for 10 min. The membranes were probed overnight with a primary rabbit antiserum against TRPM4 (1:1,000, Alomone) at 4°C and then washed three times for 10 min. The membranes were then incubated with horseradish peroxidase-labeled goat anti-rabbit secondary antiserum (1:1,000) (Thermo Fisher Scientific) for 1 h at room temperature.
Immunoreactive protein bands were detected with an iBright western blot imaging system (Thermo Fisher Scientific) using enhanced chemiluminescence reagents (Thermo Fisher Scientific; ratio of reagents A to B = 1:1). The same membrane was then stripped and probed with mouse primary antiserum against α-tubulin (1:1,000) (Thermo Fisher Scientific) to normalize the blots.
For immunocytochemistry, the HEK293 cells were seeded onto glass coverslips in 24-well plates. Once the cells reached 70%–80% confluency, transfection was conducted with TRPM4-IRES-GFP or TRPM4-HA-IRES-GFP. On the day of the experiment, the coverslips were washed in Dulbecco's phosphate-buffered saline (DPBS) and then fixed with 4% formaldehyde in PBS for 20 min at room temperature, followed by three washes in DPBS. To permeabilize the cells, the coverslips were incubated in 0.5% Triton X-100 in DPBS for 15 min. This step facilitates the removal of lipids from plasma and nuclear membranes, allowing the antibody to penetrate the cell and reach its interior. After extensive washing in DPBS, both permeable and non-permeable cells were incubated in a blocking solution (3% serum in DPBS) for 1 h. The cells were then further incubated with an anti-HA-tag monoclonal antibody (Invitrogen) diluted 1:500 for 1 h. After washing three times in DPBS, the coverslips were incubated with 1:325 diluted goat anti-mouse IgG H&L secondary antibody (Abcam) for 30 min. Then cells were washed in DPBS, the coverslips mounted with Fluorescence Mounting Medium (Dako), and then dried for 2 h in the dark. Images were captured using a confocal microscope equipped with a 405/488/594 nm laser for blue, green, and red fluorescent proteins (FLuoView; Olympus). The background intensity was normalized, and the images were analyzed using FLuoView software.
To determine the surface levels of TRPM4-HA and the YF mutants, HEK293 cells were seeded onto glass coverslips in 24-well plates. After 24 h, the cells were transfected with various types of TRPM4 cDNAs encoding IRES-GFP vector. The plasma membrane marker vector DsRed-Mem (RFP; Takara Korea Biomedical, Inc.) was used for co-transfection. Following transfection, the cells were incubated for a further 48–72 h and washed twice with PBS. The cells were fixed with 4% formaldehyde in PBS for 20 min at room temperature and then washed extensively. The cells were incubated with 4’,6’-diamidino-2-phenylindole (DAPI) (1:10,000, Thermo Fisher Scientific) solution for 5 min. After removal of the staining solution, the cells were washed three times with DPBS. Confocal fluorescence images were obtained, and the background intensity was normalized. Images were analyzed using FLuoView software.
For the electrophysiology experiments, a bath solution (140 mM NaCl, 5 mM CsCl, 1 mM CaCl2, 1 MgCl2, 10 mM glucose, and 10 mM HEPES, pH adjusted to 7.4 with NaOH) was used. A pipette solution (140 mM CsCl, 5 mM NaCl, 0.83 mM CaCl2, 1 mM MgCl2, and 1 mM EGTA, pH adjusted to 7.2 with CsOH) was used. For macro electrophysiology experiments, Bath 1 solution was prepared containing 140 mM NaCl, 5 mM KCl, 1 mM MgCl₂, 10 mM glucose, and 10 mM HEPES (pH adjusted to 7.2 with NaOH), Bath 2 solution containing 140 mM NaCl, 5 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2, 10 mM glucose, and 10 mM HEPES (pH adjusted to 7.2 with NaOH), and Bath 3 solution containing 140 mM NMDG, 5 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2, 10 mM glucose, and 10 mM HEPES (pH adjusted to 7.2 with HCL). The solutions were applied using a gravity-driven local perfusion system. All the recordings were performed at room temperature.
Patch pipettes were prepared from borosilicate glass capillaries (Warner Instruments). Whole-cell or inside-out currents were recorded using a patch-clamp amplifier (Axopatch 200B; Molecular Devices). Current–voltage (I–V) relationships were measured by applying a linear ramp protocol of 400 ms from –100 to +100 mV. The holding potential was set to approximately –40 mV, which is similar to the resting membrane potential of most HEK293 cells. The sampling interval was 200 µs, and the data were filtered at 2 kHz. The currents were analyzed using Clampfit software (Molecular Devices). Currents at ± 100 mV were used to measure the current densities by subtracting the amplitude of the initial currents from the activated TRPM4 current and dividing it by the cell capacitance (pA/pF).
Currents in either whole-cell or inside-out configurations were completely desensitized within tens of seconds and then stabilized at the basal current (BC) level. After basal currents lasting for at least 1 min, 9-phenanthrol was applied for 3 min and washed. A Na+-free solution was prepared by substituting Na+ ions with NMDG. To determine the percentage of inhibition by 9-phenanthrol, the ratio between currents at +100 mV in TRPM4 and TRPM4-HA or TRPM4-HA-YF mutants was used.
For the wound-healing assay (WHA), HEK293 cells were cultured in 24-well plates and subsequently transfected with TRPM4-HA or TRPM4-HA-YF mutant cDNA. When the cells reached 90% confluency, a small scratch was created across the cell layer using a sterile 10 µl pipette tip. Images of the cells were captured at 0, 16, 24, and 40 h with a microscope (Carl Zeiss) to assess cell proliferation and migration.
TRPM4-HA or its YF mutant was cloned into pBiFC-N-terminal VN (1–172) and PTPN6 into pBiFC-C-VC (173–273) vector. HEK293 cells were cotransfected with the two cloned BiFC vectors and incubated for 24 h. Following incubation, the cells were fixed with 4% paraformaldehyde for 20 min at room temperature and stained with DAPI (1:10,000, Thermo Fisher Scientific) for 5 min at room temperature. Coverslips were mounted with Fluorescence Mounting Medium (Dako), and Venus fluorescence signals were detected using a confocal microscope (Nikon A1).
To assess the surface expression of TRPM4-HA, Western blotting and confocal microscopy with image analysis were conducted, as illustrated in Fig. 1. As anticipated, TRPM4-HA exhibited predominant expression in the membrane fraction, similar to the wild-type TRPM4 (WT). The expression of both WT and TRPM4-HA decreased when PTPN6 was silenced using its specific shRNA (PTPN6-shRNA in Fig. 1B), indicating that the trafficking and expression of TRPM4-HA, like WT, rely on PTPN6. In Fig. 1C, the presence of HA in the intact membrane confirmed successful exposure of the HA tag inserted in the extracellular loop, enabling binding with its specific antibody and fluorescent dye in the extracellular space.
Supplementary Fig. 1 demonstrates that exogenous TRPM4 transfection increased the BC density at +100 mV by approximately three-fold, from 54.6 ± 11.0 pA/pF (GFP, n = 10) to 157.4 ± 18.83 pA/pF (TRPM4, n = 9), both in whole-cell configurations and excised inside-out patches (Supplementary Fig. 1B). Although variations in current density amplitude were observed due to differences in transfection efficiency, the response rates to chemicals remained relatively consistent, regardless of the amplitude variations (see Fig. 2 and Supplementary Fig. 1).
During recordings lasting approximately 20 min, the current levels exhibited a continuous decline (Fig. 2A). The current-voltage relationship displayed typical outward rectification for both TRPM4 (WT) and TRPM4-HA. In contrast, control cells exhibited a linear I-V relationship (Fig. 2B, C). The currents rapidly desensitized within a minute to a steady lower level, referred to as the BC, which was used as a scale for comparison with the WT.
Whole-cell currents recorded from cells transfected with TRPM4-HA were significantly inhibited by 9-phenanthrol (50.6 ± 10.2%, n = 7) at a similar rate as TRPM4-transfected cells (40.9 ± 5.7%, n = 8). In contrast, BC inhibition by 9-phenanthrol was insignificant in the GFP-transfected control cells (9.9 ± 4.6%, n = 10, see Fig. 2B).
Similar inhibition rates were observed in the inside-out configuration (57.3 ± 2.5% for TRPM4-HA, 58.6 ± 3.3% for WT). As shown in Fig. 2B, 9-phenanthrol had minimal inhibitory effects on the BC in GFP-transfected cells (Fig. 2C). When NMDG was applied instead of Na+, all the BCs obtained from cells expressing either WT or TRPM4-HA were reduced to their lowest levels (Supplementary Fig. 1C). Furthermore, when NMDG was applied to the bath solution (internal application in the excised inside-out configuration), the reductions in BCs were more pronounced at +100 mV (< 40%) compared to –100 mV (61%–79%) for both WT and TRPM4-HA. These findings confirm that the electrical properties of TRPM4-HA and WT were identical.
Previous studies have reported that TRPM4 promotes cell proliferation and migration [34]. These functions were observed in cells expressing TRPM4-HA, as shown in Fig. 3. The evaluation time was set to 24 h after transfection, as depicted in Supplementary Fig. 2. Additionally, we investigated whether the knockdown of PTPN6 using specific shRNA (PTPN6-shRNA) affected the surface expression of TRPM4-HA (Supplementary Fig. 3) and suppressed cell proliferation (Fig. 3).
The proliferation rate, assessed by WHA, was found to be higher in groups transfected with WT (54.0 ± 4.1%, n = 6) and TRPM4-HA (52.0 ± 1.3%, n = 6) compared to the GFP control group (34.5 ± 2.2%, n = 6) (Fig. 3). However, when PTPN6 was silenced, indicated by the solid bar (PTPN6-shRNA), the proliferation rates at 24 h were reduced by approximately 31.7 ± 0.9% (WT, n = 6) and 35.5 ± 2.3% (TRPM4-HA, n = 7), similar to the control GFP group (Fig. 3B). These results support the idea that TRPM4-HA retains the functional properties of WT. Furthermore, the trafficking of TRPM4-HA was found to be dependent on PTPN6, similar to WT [23].
The aforementioned results demonstrated that the properties of TRPM4 were unaffected by the addition of the HA tag, even when PTPN6 was knocked down. Since PTPN6 is necessary for the trafficking of TRPM4, this study aimed to identify the specific tyrosine (Y) residue in the N-terminus responsible for PTPN6 binding, based on previous findings indicating the exclusive binding site for PTPN6 is the N-terminus [23]. Four Y residues, namely Y75, Y256, Y393, and Y519, predicted to be involved in tyrosine phosphorylation, were selected and mutated to phenylalanines (F) using TRPM4-HA as the framework.
Initially, the expression of the four YF mutants (Y75F, Y256F, Y393F, and Y519F) in the membrane was examined. All four YF mutants were expressed in the membrane, as shown in Fig. 4A and Supplementary Fig. 3. The Y256F mutant exhibited a slightly lower expression tendency compared to the other YF mutants (∇ in Fig. 4), although this difference was not statistically significant. PTPN6 knockdown using a specific shRNA increased the retention of the mutants in the cytosolic fraction by more than two-fold, similar to the control TRPM4-HA (Fig. 4B). To confirm that the tested Y residues did not interact with PTPN6, a BiFC assay was performed. All four YF mutants interacted with PTPN6, similar to TRPM4-HA, without any statistically significant differences between the WT and mutants (Supplementary Fig. 4).
To investigate whether the TRPM4-HA-YF mutations affected the electrical properties of TRPM4, current densities were measured in cells expressing TRPM4-HA and TRPM4-HA-YF mutants. The current densities ranged from 180 to 266 pA/pF (n = 7–13) for both TRPM4-HA and TRPM4-HA-YF mutants (Fig. 5A), while the current densities in the inside-out patch configuration were in the range of 10–21 pA/pF (n = 6–16) (Fig. 5B). However, the inhibition rate of 9-phenanthrol in the TRPM4-HA-YF mutants was approximately half of that in TRPM4-HA, ranging from 22% to 45% (n = 8–13) (Fig. 5). In the inside-out patch configuration, the responses to 9-phenanthrol were also more resistant, ranging from 21% to 33% (n = 9–16), compared to TRPM4-HA (57.3 ± 2.5%, n = 6). Among the TRPM4-HA-YF mutants, the Y256F mutation exhibited the most significant decrease in the inhibition rate (summary bar charts in the right panel, Fig. 5A: 21.3 ± 4.6%, n = 12; Fig. 5B: 20.8 ± 3.9%, n = 9).
As depicted in the summary in Fig. 6, the results obtained from the WHA assay demonstrated that the TRPM4-HA-YF mutants significantly boosted cell proliferation within the range of 72.6% to 79.7% (n = 8 for each mutant) compared to the negative control (N/C). However, TRPM4-HA exhibited the highest level of proliferation at 90.1%. In contrast, the N/C group displayed only half the proliferation rate (51.6 ± 2.5%, n = 8) compared to both the TRPM4-HA-YF mutants and TRPM4-HA. When compared to non-treated naive cells (51.6 ± 2.5%, n = 8, N/C), cells with TRPM4-HA-YF mutations showed increased cell proliferation and migration ranging from 72.6 ± 3.0% to 79.7 ± 4.0% (n = 8 for each TRPM4-HA-YF mutation). However, these values were lower than those observed in the TRPM4-HA group (90.1 ± 2.8%, n = 8) (Fig. 6A). Under conditions of PTPN6 knockdown, the wound closure rates for all groups were delayed by half (50.2%–53.6%, n = 8 each) (Fig. 6B). These results indicate that the four TRPM4-HA-YF mutations are unlikely to directly participate in cell proliferation and migration.
The primary focus of the TRPM4 channel investigations has been on changes in its expression and membrane translocation. To address this, we developed a mutant variant, TRPM4-HA, which allows for the monitoring of TRPM4 trafficking in intact cells. Our aim was to assess whether its properties are comparable to those of the native TRPM4 and whether it can be utilized effectively and safely, similar to TRPM4.
Using the recently reported structure of TRPM4 [24,35], we constructed the TRPM4-HA mutant by incorporating an HA epitope. This involved inserting additional sequences before the endogenous YA located in the extracellular loop extending outside the cell. This approach preserves the integrity of the membrane, mimicking physiological conditions and minimizing the risk of overestimation or misinterpretation due to contaminants in the cytoplasmic and membrane fractions.
Detection of the epitope in the intact membrane confirmed the stable presence of the HA tag extending outward from the cell, indicating successful trafficking and surface expression of TRPM4-HA. Importantly, TRPM4-HA exhibited channel properties identical to TRPM4, as demonstrated by electrophysiology and pharmacology experiments. Furthermore, the cell proliferative function and dependence on PTPN6 for trafficking were conserved with this mutant, mirroring those of TRPM4. These results establish that TRPM4-HA functions as a TRPM4 channel with an extracellular HA epitope.
Investigations using TRPM4-HA and its YF mutants did not provide clear information about the binding site with PTPN6, which is crucial for TRPM4 trafficking. Interestingly, the TRPM4-HA-YF mutants showed reduced sensitivity to 9-phenanthrol compared to TRPM4-HA. Among these mutants, Y256F exhibited the lowest sensitivity, suggesting that Y256 is likely part of the 9-phenanthrol binding site. Y256 is located in the α6 helix, which forms part of the bottom of the N-terminal TRPM4 homology regions 1 and 2 (MHR1/2). These regions are positioned beneath the C-terminal domain and may play a role in gating and ion channel activities [24]. Therefore, Y256 within MHR1/2 is a potential site for 9-phenanthrol to modulate ion channel activity. Additionally, the Y256F mutant appeared to resist membrane translocation, possibly contributing to lower channel currents. However, since the current sizes showed little variation, it is more likely that the studied Y residues all play a role in modulating channel function.
BiFC analysis targeting the four residues predicted by tyrosine phosphorylation did not provide evidence of protein-protein interactions between PTPN6 and each YF mutant. This suggests that PTPN6 stably binds to the non-phosphorylated form of TRPM4. However, this study examining the selected Y residues could not determine whether PTPN6 directly dephosphorylates TRPM4 or identify the specific site(s) responsible for this effect. The examination under PTPN6 knockout conditions in this study did not yield direct information about PTPN6's effect on tyrosine phosphorylation.
In conclusion, TRPM4-HA fulfills the requirements for investigating TRPM4, serving as a valuable tool for identifying critical structures involved in pathway expression and migration, thanks to the internal HA tag in the extracellular loop. TRPM4-HA is particularly well-suited for studying cell functions related to morphological changes and expression patterns in relation to TRPM4.
Supplementary data including four figures can be found with this article online at https://doi.org/10.4196/kjpp.2023.27.4.417.
ACKNOWLEDGEMENTS
We thank Professor Jae Yong Park (Korea University college of Health Science) for technical advices on BiFC analysis.
REFERENCES
1. Launay P, Fleig A, Perraud AL, Scharenberg AM, Penner R, Kinet JP. 2002; TRPM4 is a Ca2+-activated nonselective cation channel mediating cell membrane depolarization. Cell. 109:397–407. DOI: 10.1016/S0092-8674(02)00719-5. PMID: 12015988.

2. Nilius B, Prenen J, Droogmans G, Voets T, Vennekens R, Freichel M, Wissenbach U, Flockerzi V. 2003; Voltage dependence of the Ca2+-activated cation channel TRPM4. J Biol Chem. 278:30813–30820. DOI: 10.1074/jbc.M305127200. PMID: 12799367.

3. Murakami M, Xu F, Miyoshi I, Sato E, Ono K, Iijima T. 2003; Identification and characterization of the murine TRPM4 channel. Biochem Biophys Res Commun. 307:522–528. DOI: 10.1016/S0006-291X(03)01186-0. PMID: 12893253.

4. Jang Y, Lee Y, Kim SM, Yang YD, Jung J, Oh U. 2012; Quantitative analysis of TRP channel genes in mouse organs. Arch Pharm Res. 35:1823–1830. DOI: 10.1007/s12272-012-1016-8. PMID: 23139135.

5. Guinamard R, Bouvagnet P, Hof T, Liu H, Simard C, Sallé L. 2015; TRPM4 in cardiac electrical activity. Cardiovasc Res. 108:21–30. DOI: 10.1093/cvr/cvv213. PMID: 26272755.

6. Wang H, Xu Z, Lee BH, Vu S, Hu L, Lee M, Bu D, Cao X, Hwang S, Yang Y, Zheng J, Lin Z. 2019; Gain-of-function mutations in TRPM4 activation gate cause progressive symmetric erythrokeratodermia. J Invest Dermatol. 139:1089–1097. DOI: 10.1016/j.jid.2018.10.044. PMID: 30528822.

7. Çoban G, Yildiz P, Doğan B, Şahin N, Gücin Z. 2021; Expression of transient receptor potential melastatin 4 in differential diagnosis of eosinophilic renal tumors. Mol Clin Oncol. 15:230. DOI: 10.3892/mco.2021.2393. PMID: 34631055. PMCID: PMC8461610.

8. Kappel S, Stokłosa P, Hauert B, Ross-Kaschitza D, Borgström A, Baur R, Galván JA, Zlobec I, Peinelt C. 2019; TRPM4 is highly expressed in human colorectal tumor buds and contributes to proliferation, cell cycle, and invasion of colorectal cancer cells. Mol Oncol. 13:2393–2405. DOI: 10.1002/1878-0261.12566. PMID: 31441200. PMCID: PMC6822246. PMID: 1593c9ef94da47ea9b0e9396bb130ad4.

9. Sagredo AI, Sagredo EA, Cappelli C, Báez P, Andaur RE, Blanco C, Tapia JC, Echeverría C, Cerda O, Stutzin A, Simon F, Marcelain K, Armisén R. 2018; TRPM4 regulates Akt/GSK3-β activity and enhances β-catenin signaling and cell proliferation in prostate cancer cells. Mol Oncol. 12:151–165. DOI: 10.1002/1878-0261.12100. PMID: 28614631. PMCID: PMC5792731. PMID: 39b1d5fb4e064e63b1ddea25c367a8f3.

10. Cáceres M, Ortiz L, Recabarren T, Romero A, Colombo A, Leiva-Salcedo E, Varela D, Rivas J, Silva I, Morales D, Campusano C, Almarza O, Simon F, Toledo H, Park KS, Trimmer JS, Cerda O. 2015; TRPM4 is a novel component of the adhesome required for focal adhesion disassembly, migration and contractility. PLoS One. 10:e0130540. DOI: 10.1371/journal.pone.0130540. PMID: 26110647. PMCID: PMC4482413. PMID: 14e94f17630944cca89bcbf1c358b2f2.

11. Launay P, Cheng H, ivatsan S Sr, Penner R, Fleig A, Kinet JP. 2004; TRPM4 regulates calcium oscillations after T cell activation. Science. 306:1374–1377. DOI: 10.1126/science.1098845. PMID: 15550671.

12. Vennekens R, Olausson J, Meissner M, Bloch W, Mathar I, Philipp SE, Schmitz F, Weissgerber P, Nilius B, Flockerzi V, Freichel M. 2007; Increased IgE-dependent mast cell activation and anaphylactic responses in mice lacking the calcium-activated nonselective cation channel TRPM4. Nat Immunol. 8:312–320. DOI: 10.1038/ni1441. PMID: 17293867.

13. Barbet G, Demion M, Moura IC, Serafini N, Léger T, Vrtovsnik F, Monteiro RC, Guinamard R, Kinet JP, Launay P. 2008; The calcium-activated nonselective cation channel TRPM4 is essential for the migration but not the maturation of dendritic cells. Nat Immunol. 9:1148–1156. DOI: 10.1038/ni.1648. PMID: 18758465. PMCID: PMC2956271.

14. Shimizu T, Owsianik G, Freichel M, Flockerzi V, Nilius B, Vennekens R. 2009; TRPM4 regulates migration of mast cells in mice. Cell Calcium. 45:226–232. DOI: 10.1016/j.ceca.2008.10.005. PMID: 19046767.
15. Gao Y, Liao P. 2019; TRPM4 channel and cancer. Cancer Lett. 454:66–69. DOI: 10.1016/j.canlet.2019.04.012. PMID: 30980865.

16. Watanabe H, Murakami M, Ohba T, Ono K, Ito H. 2009; The pathological role of transient receptor potential channels in heart disease. Circ J. 73:419–427. DOI: 10.1253/circj.CJ-08-1153. PMID: 19202304.

17. Wang C, Naruse K, Takahashi K. 2018; Role of the TRPM4 channel in cardiovascular physiology and pathophysiology. Cells. 7:62. DOI: 10.3390/cells7060062. PMID: 29914130. PMCID: PMC6025450. PMID: 1bba7c5b29624e5592a2e292f89d525c.

18. Otsuka Saito K, Fujita F, Toriyama M, Utami RA, Guo Z, Murakami M, Kato H, Suzuki Y, Okada F, Tominaga M, Ishii KJ. 2023; Roles of TRPM4 in immune responses in keratinocytes and identification of a novel TRPM4-activating agent. Biochem Biophys Res Commun. 654:1–9. DOI: 10.1016/j.bbrc.2023.02.062. PMID: 36871485.

19. Holzmann C, Kappel S, Kilch T, Jochum MM, Urban SK, Jung V, Stöckle M, Rother K, Greiner M, Peinelt C. 2015; Transient receptor potential melastatin 4 channel contributes to migration of androgen-insensitive prostate cancer cells. Oncotarget. 6:41783–41793. DOI: 10.18632/oncotarget.6157. PMID: 26496025. PMCID: PMC4747188.

20. Berg KD, Soldini D, Jung M, Dietrich D, Stephan C, Jung K, Dietel M, Vainer B, Kristiansen G. 2016; TRPM4 protein expression in prostate cancer: a novel tissue biomarker associated with risk of biochemical recurrence following radical prostatectomy. Virchows Arch. 468:345–355. DOI: 10.1007/s00428-015-1880-y. PMID: 26590985.

21. Sagredo AI, Sagredo EA, Pola V, Echeverría C, Andaur R, Michea L, Stutzin A, Simon F, Marcelain K, Armisén R. 2019; TRPM4 channel is involved in regulating epithelial to mesenchymal transition, migration, and invasion of prostate cancer cell lines. J Cell Physiol. 234:2037–2050. DOI: 10.1002/jcp.27371. PMID: 30343491.

22. Ashida S, Nakagawa H, Katagiri T, Furihata M, Iiizumi M, Anazawa Y, Tsunoda T, Takata R, Kasahara K, Miki T, Fujioka T, Shuin T, Nakamura Y. 2004; Molecular features of the transition from prostatic intraepithelial neoplasia (PIN) to prostate cancer: genome-wide gene-expression profiles of prostate cancers and PINs. Cancer Res. 64:5963–5972. DOI: 10.1158/0008-5472.CAN-04-0020. PMID: 15342375.

23. Lee DK, Park JY, Yoo JC, Byun EH, Bae YJ, Lee YS, Park N, Kang D, Han J, Park JY, Hwang E, Hong SG. 2018; PTPN6 regulates the cell-surface expression of TRPM4 channels in HEK293 cells. Pflugers Arch. 470:1449–1458. DOI: 10.1007/s00424-018-2161-9. PMID: 29931651.

24. Winkler PA, Huang Y, Sun W, Du J, Lü W. 2017; Electron cryo-microscopy structure of a human TRPM4 channel. Nature. 552:200–204. DOI: 10.1038/nature24674. PMID: 29211723.

25. Guinamard R, Hof T, Del Negro CA. 2014; The TRPM4 channel inhibitor 9-phenanthrol. Br J Pharmacol. 171:1600–1613. DOI: 10.1111/bph.12582. PMID: 24433510. PMCID: PMC3966741.

26. Jarvik JW, Telmer CA. 1998; Epitope tagging. Annu Rev Genet. 32:601–618. DOI: 10.1146/annurev.genet.32.1.601. PMID: 9928493.

27. Maue RA. 2007; Understanding ion channel biology using epitope tags: progress, pitfalls, and promise. J Cell Physiol. 213:618–625. DOI: 10.1002/jcp.21259. PMID: 17849449.
28. Kennedy ME, Nemec J, Clapham DE. 1996; Localization and interaction of epitope-tagged GIRK1 and CIR inward rectifier K+ channel subunits. Neuropharmacology. 35:831–839. DOI: 10.1016/0028-3908(96)00132-3. PMID: 8938714.

29. Peters KW, Qi J, Johnson JP, Watkins SC, Frizzell RA. 2001; Role of snare proteins in CFTR and ENaC trafficking. Pflugers Arch. 443 Suppl 1:S65–69. DOI: 10.1007/s004240100647. PMID: 11845306.

30. Czech MP, Chawla A, Woon CW, Buxton J, Armoni M, Tang W, Joly M, Corvera S. 1993; Exofacial epitope-tagged glucose transporter chimeras reveal COOH-terminal sequences governing cellular localization. J Cell Biol. 123:127–135. DOI: 10.1083/jcb.123.1.127. PMID: 8408193. PMCID: PMC2119811.

31. Wadzinski BE, Eisfelder BJ, Peruski LF Jr, Mumby MC, Johnson GL. 1992; NH2-terminal modification of the phosphatase 2A catalytic subunit allows functional expression in mammalian cells. J Biol Chem. 267:16883–16888. DOI: 10.1016/S0021-9258(18)41867-4. PMID: 1380955.

32. Kast C, Canfield V, Levenson R, Gros P. 1996; Transmembrane organization of mouse P-glycoprotein determined by epitope insertion and immunofluorescence. J Biol Chem. 271:9240–9248. DOI: 10.1074/jbc.271.16.9240. PMID: 8621583.

33. Poteser M, Graziani A, Rosker C, Eder P, Derler I, Kahr H, Zhu MX, Romanin C, Groschner K. 2006; TRPC3 and TRPC4 associate to form a redox-sensitive cation channel. Evidence for expression of native TRPC3-TRPC4 heteromeric channels in endothelial cells. J Biol Chem. 281:13588–13595. DOI: 10.1074/jbc.M512205200. PMID: 16537542.
34. Armisén R, Marcelain K, Simon F, Tapia JC, Toro J, Quest AF, Stutzin A. 2011; TRPM4 enhances cell proliferation through up-regulation of the β-catenin signaling pathway. J Cell Physiol. 226:103–109. DOI: 10.1002/jcp.22310. PMID: 20625999.

35. Autzen HE, Myasnikov AG, Campbell MG, Asarnow D, Julius D, Cheng Y. 2018; Structure of the human TRPM4 ion channel in a lipid nanodisc. Science. 359:228–232. DOI: 10.1126/science.aar4510. PMID: 29217581. PMCID: PMC5898196.

Fig. 1
Expression of HA-tagged TRPM4 in the intact membrane.
(A) Schematic illustration of TRPM4 with a HA epitope (nine amino acids) inserted in the extracellular loop between the fifth (S5) and sixth (S6) transmembrane domains. (B) Western blot of the expression of the HA-tagged TRPM4 channel, TRPM4-HA, in the cell membrane and determination of its PTPN6 dependency. The membrane expression of both TRPM4 and TRPM4-HA were disrupted by silencing PTPN6 (PTPN6 shRNA). The data were obtained from three replicate experiments. (C) Fluorescence confocal microscopy image of cells labeled with anti-HA antibody and Alexa 594. GFP images (left) of cells expressing IRES-GFP vector containing TRPM4-HA, captured at a wavelength of 488 nm. The red fluorescent images (right) were also captured at a wavelength of 488 nm. The top and middle images are of permeabilized cells and the lower images are of non-treated cells with intact membranes. Asterisks indicate significant differences from the control group: *p < 0.05, ***p < 0.001; scale bar represents 10 μm, and all data are shown as means ± SEM. HA, hemagglutinin; TRPM4, transient receptor potential melastatin 4; PTPN6, protein tyrosine phosphatase, non-receptor type 6; WT, wild-type.
Fig. 2
The channel properties of TRPM4-HA were consistent with those of TRPM4 (WT).
(A) A representative sequence from the electrophysiological experiments. Currents were elicited using ramp pulses every 5 sec in the presence of 100 μM Ca2+. A bathing solution containing 9-phenantrol or NMDG was applied following stable basal currents observed for at least 1 min post-desensitization. The figure shows representative tracings obtained in the excised inside-out configuration. (B) The I-V relationships affected by 9-phenanthrol are presented as bar charts. Representative I-V current relationships from cells transfected with GFP only as a control (left, white), WT (middle, gray), and TRPM4-HA (right, dark gray) are shown. The rates of inhibition by 9-phenanthrol are summarized, with error bars on the right. (C) Similar to (B), the I-V relationship and data summaries from the excised inside-out configuration are shown on the right. The recording configurations are represented as cartoons on the left side of each panel. The black and red current traces were recorded immediately before (control) and 3 min after applying 9-phenanthrol, respectively. Asterisks indicate significant differences between the data before and after applying 9-phenanthrol: **p < 0.01, ***p < 0.001. All data are presented as means ± SEM. HA, hemagglutinin; TRPM4, transient receptor potential melastatin 4; NMDG, N-Methyl-D-glucamine; WT, wild-type; NS, not significant.
Fig. 3
The TRPM4-HA was found to regulate cell proliferation and migration, and its activity was dependent on PTPN6.
(A) Representative images of the wound-healing assay (n = 6) with cells overexpressing TRPM4 (WT) and TRPM4-HA, and a summary bar chart is presented in (B). The rate of wound closure 24 h post-scratching was assessed in six replicate experiments. Solid lines indicate images and data from cells with silencing of PTPN6 expression. Statistical analysis revealed significant differences compared with the control, with **p < 0.01, ****p < 0.0001. All data are presented as means ± SEM. HA, hemagglutinin; TRPM4, transient receptor potential melastatin 4; PTPN6, protein tyrosine phosphatase, non-receptor type 6; WT, wild-type.
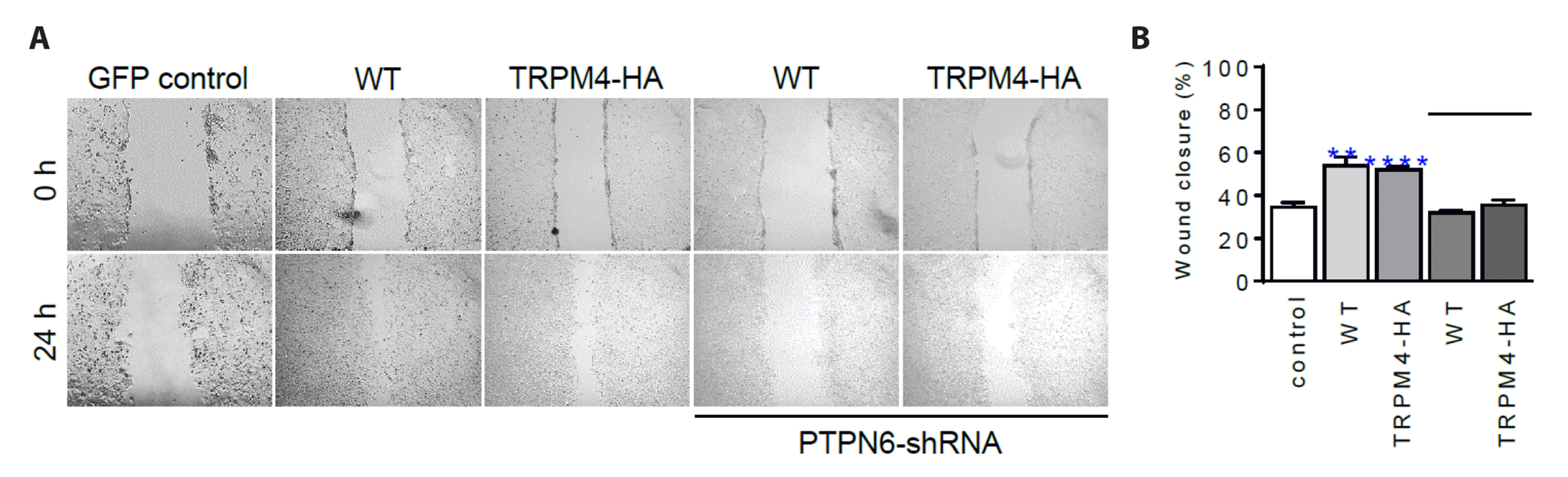
Fig. 4
The surface expression of the YF mutants is not independent of PTPN6.
(A) Western blot analysis clearly showing that both TRPM4-HA as a control and TRPM4-HA-YF mutants were expressed at similar levels (0.90 ± 0.03–1.16 ± 0.04, n = 3) on the cell surface, compared to the levels in the cytosolic fraction (0.13 ± 0.06–0.20 ± 0.11, n = 3). (B) In repeated analysis under PTPN6 knockdown conditions to interfere with channel trafficking, silencing PTPN6 greatly increased the levels of all TRPM4-HA-YF mutants in the cytosolic fraction (0.35 ± 0.11–0.48 ± 0.10, n = 3). The Y256F (∇) mutant exhibited a degree of resistance to trafficking, although the difference was not statistically significant. The Western blot images are representative of three replicate experiments. Statistical analysis revealed significant differences compared with the control, with *p < 0.05, **p < 0.01. All data are presented as means ± SEM. HA, hemagglutinin; TRPM4, transient receptor potential melastatin 4; PTPN6, protein tyrosine phosphatase, non-receptor type 6.
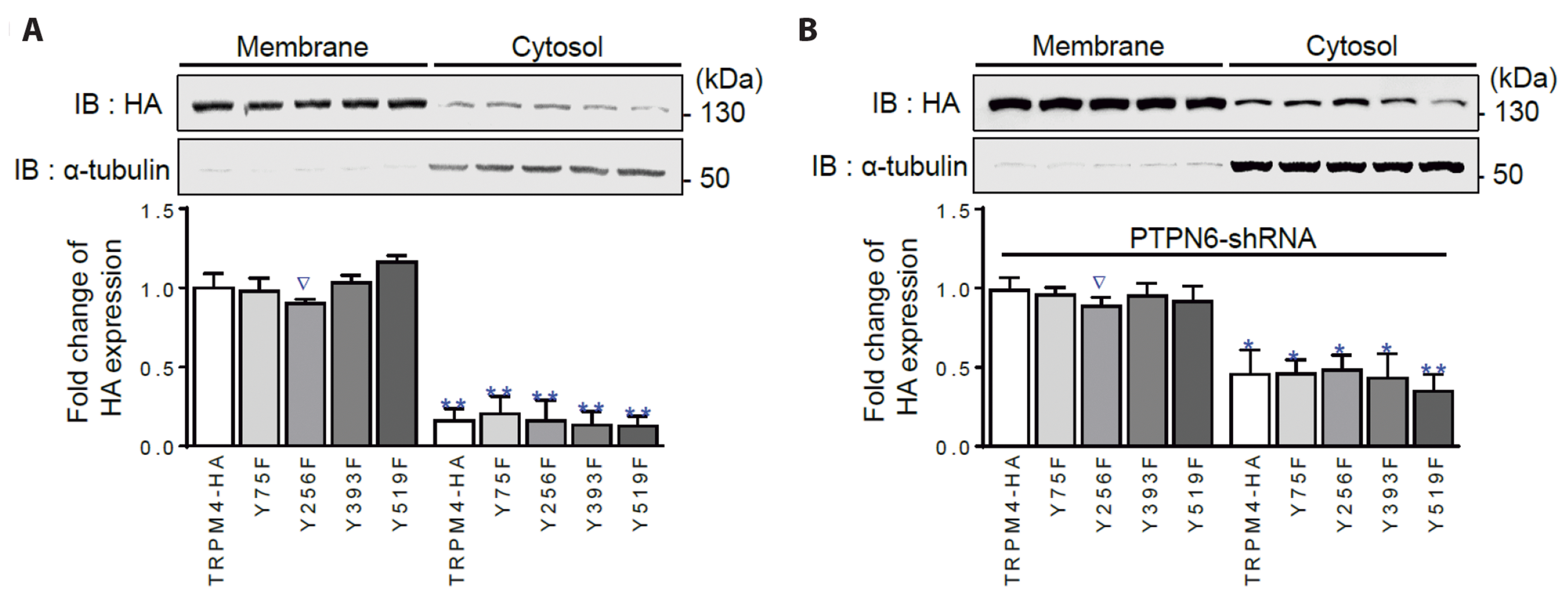
Fig. 5
Channel currents in TRPM4-HA-YF mutants were found to be resistant to 9-phenanthrol.
(A) The I-V relationships for whole-cell currents of the TRPM4-HA-YF mutants were lower than that of TRPM4-HA, although not statistically significant, except for Y256F (p = 0.003, n = 12). (B) The I-V relationships of the excised inside-out currents in the TRPM4-HA-YF mutants were also evaluated. In isolated membrane patches without cytosol, the currents in cells with the TRPM4-HA-YF mutants were found to be more resistant to inhibition by 9-phenanthrol, with inhibition rates ranging from approximately 25% to half the level of inhibition observed in TRPM4-HA (57.3 ± 2.5%, n = 6). The black and red current traces were recorded immediately before (control) and 3 min after applying 9-phenanthrol, respectively. Bar charts were obtained from currents measured at +100 mV. Asterisks indicate significant differences between data before and after the application of 9-phenanthrol, **p < 0.01, ***p < 0.001. ****p < 0.0001. All data are shown as means ± SEM. HA, hemagglutinin; TRPM4, transient receptor potential melastatin 4.
Fig. 6
Effects of TRPM4-HA-YF mutation on cell proliferation and migration and the dependence on PTPN6.
(A) Representative images of cell proliferation and migration assessed by wound-healing assay (WHA) with naive (N/C), TRPM4-HA, and TRPM4-HA-YF mutant-overexpressing cells. TRPM4-HA-YF mutations resulted in a significant reduction in cell proliferation and migration by approximately 30% or more, compared with the inhibition rate of 10% observed for TRPM4-HA. (B) In replicate experiments, cells transfected with TRPM4-HA with scrambled RNA or with TRPM4-HA-YF mutants and PTPN6 shRNA were subjected to WHA. Under PTPN6 knockdown conditions, wound closure was delayed by approximately 50% in the cases indicated by a solid bar. Wound closure was assessed 24 h post-scratching in replicate experiments (n = 8). Solid lines indicate images and data from PTPN6 knockdown cells. *p < 0.05, **p < 0.01, ***p < 0.001 compared to the control. All data are presented as means ± SEM. HA, hemagglutinin; TRPM4, transient receptor potential melastatin 4; PTPN6, protein tyrosine phosphatase, non-receptor type 6; N/C, negative control.





 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download