Abstract
Crohn's disease (CD) is a chronic inflammatory illness of the digestive system with unknown etiology, and its incidence is increasing worldwide. However, there are currently no effective treatments or medications available for individuals with CD. Therefore, novel therapeutic strategies are urgently needed. The bioactive compounds and targets associated with compounds of Qinghua Xiaoyong Formula (QHXYF) were examined using The Traditional Chinese Medicine Systems Pharmacology database, and 5 disease target databases were also used to identify CD-related disease targets. A total of 166 overlapping targets were identified from QHXYF-related and CD-related disease targets and they were found to be enriched in oxidative stress-related pathways and the PI3K/AKT signaling pathway. Molecular docking was then used to predict how the bioactive compounds would bind to the hub targets. It was found that quercetin could be the core bioactive compound and had good binding affinity to the top 5 hub targets. Finally, animal experiments were performed to further validate the findings, and the results revealed that QHXYF or quercetin inhibited 2,4,6-trinitrobenzenesulfonic acid-induced inflammation and oxidative stress processes by inhibiting the PI3K/AKT pathway, thereby improving CD symptoms. These findings suggest that QHXYF and quercetin may be potential novel treatments for CD.
Crohn's disease (CD) is a chronic inflammatory illness of the digestive system with an unknown etiology, and its incidence is increasing worldwide [1]. Over the course of a patient's lifetime, CD frequently relapses and can result in complications, such as colon stenosis, abscesses, and fistulas. CD is closely related to inflammation and the induction of oxidative stress, which can worsen over time [2,3]. Currently, there are no effective treatments or medications available for individuals with CD [4]. The main goal of treatment is to alleviate colonic inflammation, control acute flares of the disease, and reduce recurrence through targeted intervention of inflammatory molecules. Therefore, novel therapeutic strategies are urgently needed.
In the treatment of CD and colitis-related diseases, traditional Chinese medicine (TCM) has shown great therapeutic potential. For example, the response rate of the Chinese herbal medicine Sophora flavescens (Kushen) in the clinical treatment of ulcerative colitis (UC) was similar to that of conventional therapies [5]. The results of basic experiments have shown that its extract can modulate the dextran sodium sulfate (DSS)-induced inflammatory response and Th17/Treg, thereby alleviating UC symptoms [6]. "Qinghua Xiaoyong Formula" (QHXYF) is a TCM formula that can be used to treat UC in clinical practice. The main components of this formula are collectively known as San Huang, which include Scutellaria baicalensis (Huangqin), Coptis chinensis (Huanglian), and Phellodendri cortex (Huangbai), which can relieve heat and dispel dampness. S. baicalensis has been shown to alleviate the symptoms of colitis in a mouse model of colitis by inhibiting inflammation and oxidative stress [7]. C. chinensis might prevent intestinal barrier damage by regulating intestinal bacteria dysbiosis and suppressing inflammatory response [8]. P. cortex also has anti-inflammatory effects, as the methanolic extract of P. cortex has been shown to attenuate lipopolysaccharide (LPS)-induced acute airway inflammation in mice [9]. However, there are limited reports on QHXYF in the treatment of colitis-related diseases such as CD, and the mechanism remains unclear.
In this study, we aimed to investigate the probable mechanism of QHXYF using network pharmacology and molecular docking, as well as evaluate the effectiveness of QHXYF and its bioactive compounds in treating CD in mice. Our research offers a fresh perspective to expand the use of TCM in conditions related to colitis.
We utilized the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, https://tcmspw.com/tcmsp.php) [10] to screen for the main bioactive compounds of each herb present in QHXYF. Our screening criteria included an oral bioavailability of ≥ 30% and drug likeness of ≥ 0.18. Additionally, we retrieved targets associated with these bioactive compounds from the TCMSP database. The UniProt database (https://www.uniprot.org/) was then used to identify the gene symbols of these compound-associated targets.
To identify CD-related disease targets, we searched several databases of disease-associated genes, including GeneCards (https://www.genecards.org/), OMIM (https://omim.org/), TTD (http://db.idrblab.net/ttd/), PharmGkb (https://www.pharmgkb.org/), and DrugBank (https://www.drugbank.com/) using "Crohn’s Disease" as a keyword, as previously reported [11]. Next, targets of QHXYF and CD-related disease targets were input into the BioLadder software (www.bioladder.cn/web/#/pro/index) to create a Venn diagram to identify the overlapping targets. These overlapping targets were identified as CD-related targets of bioactive compounds and were used for further analysis.
We first used Cytoscape 3.9.1 to construct a composition-disease-target network diagram. Then, NetworkAnalyzer tools in Cytoscape 3.9.1 was used to perform topology analysis and degree ranking.
Subsequently, Online STRING 11.5 database (https://string-db.org/) was used to predict the protein–protein interaction (PPI) of overlapping targets. PPI network was constructed and visualized using Cytoscape 3.9.1 and cytoHubba. The top 10 core nodes were identified and ranked by the maximum neighborhood component (MNC) method using the "cytoHubba" plugin in Cytoscape 3.9.1 [12].
To identify biological processes (BP) and signaling pathways enriched in CD-related and bioactive compound-targeting gene set, “clusterProfiler” package in R software was used to analyse the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The GO term analysis included BP, cellular component, and molecular function. A p-value of less than 0.05 was considered statistically significant.
The crystal structures of AKT1 (6HHF), TP53 (5O1F), TNF (2AZ5), IL6 (4CNI), and VEGFA (4QAF) were downloaded from the PDB library and introduced into the Chimera 1.16 software for protein structure construction. The SDF format of quercetin was then obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). Molecular docking analysis used to predict protein-compound binding activity was performed using an online tool CB-Dock (http://clab.labshare.cn/cb-dock/php/manual.php), which was designed to perform blind docking at predicted sites [13]. A specific binding activity between a ligand and a protein is represented by the Vina score, and the stability of the ligand's binding to the protein is reflected by a lower Vina score.
C57BL/6 mice (6 to 8-week old) were obtained from Shanghai Slake Experimental Animal Co., Ltd. The experiments were approved by the Guide for the Care and Use of Laboratory Animals and Shuguang Hospital affiliated with Shanghai University of TCM (PZSHUTCM210702005). The mice were divided into four groups (n = 6 per group): Sham, TNBS, TNBS + QHXYF (1 g/kg), and TNBS + Quercetin (30 mg/kg).
For TNBS treatment, all mice were administered 2,4,6-trinitrobenzenesulfonic acid (TNBS) at a concentration of 100 mg/kg by enema for 7 consecutive days to induce colitis. The sham group was treated with saline. Additionally, TNBS-treated mice were gavaged with 1 g/kg QHXYF (10 g S. baicalensis, 3 g C. chinensis, and 10 g P. cortex) or Quercetin (30 mg/kg) for 7 consecutive days. The sham group was administered distilled water. The body weight of the mice was monitored daily for 7 consecutive days. On day 7, the mice were euthanized, the colons were removed, photographed, and the length of colons was measured.
DAI was determined by assessing body weight, stool consistency, and blood in the stool, as previously reported [14]. In brief, weight loss, diarrhea and blood in stool were assessed as follows: (A) (weight loss) score 0, none; score 1, 1%–5%; score 2, 5%–10%; score 3, 10%–20%; score 4, > 20%; (B) (diarrhea) score 0, normal; score 2, loose stools; score 4, watery diarrhea; (C) (blood in stool) score 0, normal; score 2, slight bleeding; score 4, gross bleeding.
The colon tissues of each group of mice were removed, treated with 4% paraformaldehyde, and cut into 4 µm sections. After deparaffinization, the samples were rehydrated and stained with hematoxylin and eosin (H&E). Images were taken using a light microscope. Three observers, who were blinded to the mice’s grouping, evaluated each set of colon sections.
Total protein was extracted from the colon tissues of each group of mice as previously described [15]. Protein lysates from cells were loaded onto a 10% SDS-polyacrylamide gel and separated by electrophoresis. The proteins were then transferred to the polyvinylidene difluoride membranes (Millipore), and blocked with 5% non-fat milk for 1 h at room temperature. The membranes were then incubated with primary antibodies overnight at 4°C. Primary antibodies used in this study are as follows: anti-p-PI3K (ab278545, 1:5,000), anti-PI3K (ab32089,1:5,000), anti-p-AKT (ab38449, 1:4,500), anti-AKT (ab179463, 1:4,500) and anti-β-actin (ab8227, 1:1,000). All the antibodies were purchased from Abcam. The following day, the membranes were washed 3 times in TBST and incubated with HRP-conjugated secondary IgG antibody (ab205718, 1:2,000, Abcam) for 1 h at room temperature. Lastly, protein signals were detected using enhanced chemiluminescence reagent (Beyotime), and ImageJ was used for quantification.
Serum and colon tissues were collected from mice. Inflammation markers tumor necrosis factor-α (TNF-α), interleukin (IL)-6, and IL-1β in serum, as well as oxidative stress markers monochrome adapter (MDA), superoxide dismutase (SOD), and glutathione (GSH) in colon tissues, were quantified using corresponding ELISA kits (Beyotime) according to manufacturer’s instructions. The absorbance values were measured at 450 nm.
Kinase inhibition was assessed using the Z’ LYTE kinase assay kit (Thermo Fisher Scientific), which is a cell-free in vitro assay. This assay method employs a Fluorescence Resonance Energy Transfer-based, coupled-enzyme format and is based on the differential sensitivity of phosphorylated and non-phosphorylated peptides to proteolytic cleavage. The inhibitory activity of the top 5 proteins (AKT, TP53, TNF, IL6, and VEGFA) was measured using the Z’ LYTE kinase assay, and the percentage of inhibition of each kinase by pirfenidone was measured according to the manufacturer’s instructions.
All data are expressed as mean ± standard deviation. Each set of experiments was repeated three times. For statistical analysis, GraphPad Prism software 7.04 was used. The student's t-test was used to compare difference between the two groups, whereas comparisons between multiple groups were performed using one-way ANOVA. A p-value of less than 0.05 was considered statistically significant.
First, we screened CD-related disease genes using five disease databases, namely OMIM, GeneCards, TTD, PharmGkb, and DrugBank, and identified a total of 4,226 CD-related genes (Fig. 1A). Next, the TCMSP database was used to screen for bioactive compounds in QHXYF. QHXYF consists of S. baicalensis, C. chinensis, and P. cortex. A total of 74 bioactive compounds were identified, including 36 in S. baicalensis, 14 in C. chinensis, and 37 in P. cortex. In addition, 235 targets were identified for these 74 bioactive compounds. The Venn diagram in Fig. 1B shows that a total of 166 overlapping targets were identified from QHXYF-related and CD-related disease targets. The intersection between bioactive compound and disease targets further indicates the potential interplay between QHXYF bioactive compounds and CD (Fig. 1C). Next, key component-target topology analysis was performed using Cytoscape 3.9.1 and NetworkAnalyzer tools, and our findings showed that quercetin was the drug compound with the highest degree in QHXYF (Fig. 1D). This was followed by wogonin, baicalein, and coptisine.
To further confirm whether the 166 overlapping targets that were enriched in biological activities were associated with CD, GO and KEGG enrichment analyses were performed using R. The results showed that the functions of QHXYF and CD target genes may be related to response to oxidative stress pathway and PI3K/AKT signaling pathway (Fig. 2A, B). Subsequently, we analyzed the PPI of these 166 overlapping genes using the STRING online database, and the PPI topological network relationship diagram is shown in Fig. 2C.
Based on the PPI network, the top 10 core nodes were identified and ranked using the MNC method with "cytoHubba" plugin (Cytoscape 3.9.1). These core nodes were identified as hub genes and included AKT1, TP53, TNF, IL6, VEGFA, JUN, IL1B, CASP3, PTGS2, and MYC (Fig. 3A, B). Subsequently, we used a Sankey diagram to show the relationship between herbs, bioactive compounds, and the top 5 hub targets. We found that quercetin, which is shared by C. chinensis and P. cortex, targets all of the top 5 hub genes (Fig. 3C), suggesting that quercetin may be a key bioactive compound of QHXYF. As a result, molecular docking was performed on quercetin with AKT1, TP53, TNF, IL6, and VEGFA. Our results showed that quercetin and AKT1 had the highest cavity size and the lowest Vina score, indicating a strong binding affinity between quercetin and AKT1 (Fig. 3D, E). We also performed molecular docking of quercetin with the other top 5 hub genes, which are shown in Fig. 3F. In addition, we conducted a Z'-LYTE kinase inhibition assay to validate the binding effect of quercetin to top 5 proteins. The IC50 values of quercetin on AKT1, TP53, TNF, IL6, and VEGFA were 2.288 μM, 4.037 μM, 8.753 μM, 14.31 μM, and 19.76 μM, respectively (Supplementary Fig. 1). Our results indicated that quercetin had a strong inhibitory effect on AKT1. Therefore, we screened AKT1-related pathways for further analysis.
To explore the effect of QHXYF or quercetin on symptom of colitis, a TNBS-induced mouse model was established. We found that mice in the TNBS group exhibited a significant decrease in body weight and an increase in DAI, whereas QHXYF or quercetin treatment could alleviate the body weight loss and decreased DAI (Fig. 4A, B). Moreover, photographs of colons showed that the length of TNBS-induced colon was increased by QHXYF or quercetin treatment (Fig. 4C). Next, we used H&E staining to assess the pathological condition of the colon. As shown in Fig. 4D, TNBS resulted in extensive infiltration of inflammatory cells and severe diffuse disruption of the colonic epithelium. However, QHXYF or quercetin significantly ameliorated TNBS-induced epithelial injury. In summary, the data suggested that QHXYF or quercetin improved symptom in TNBS-induced mice.
We then examined whether QHXYF or quercetin could affect inflammation in TNBS-induced mouse model. The ELISA results verified that TNF-α, IL-6, and IL-1β levels were increased in TNBS-induced mice, and these were reversed by QHXYF or quercetin treatment (Fig. 5A). In addition, we also examined oxidative stress markers by ELISA. The level of MDA was upregulated, while the levels of SOD and GSH were downregulated in TNBS-induced mice. However, these were reversed by treatment with QHXYF or quercetin (Fig. 5B). These results indicated that QHXYF or quercetin could inhibit inflammation and oxidative stress markers in TNBS-induced mouse model.
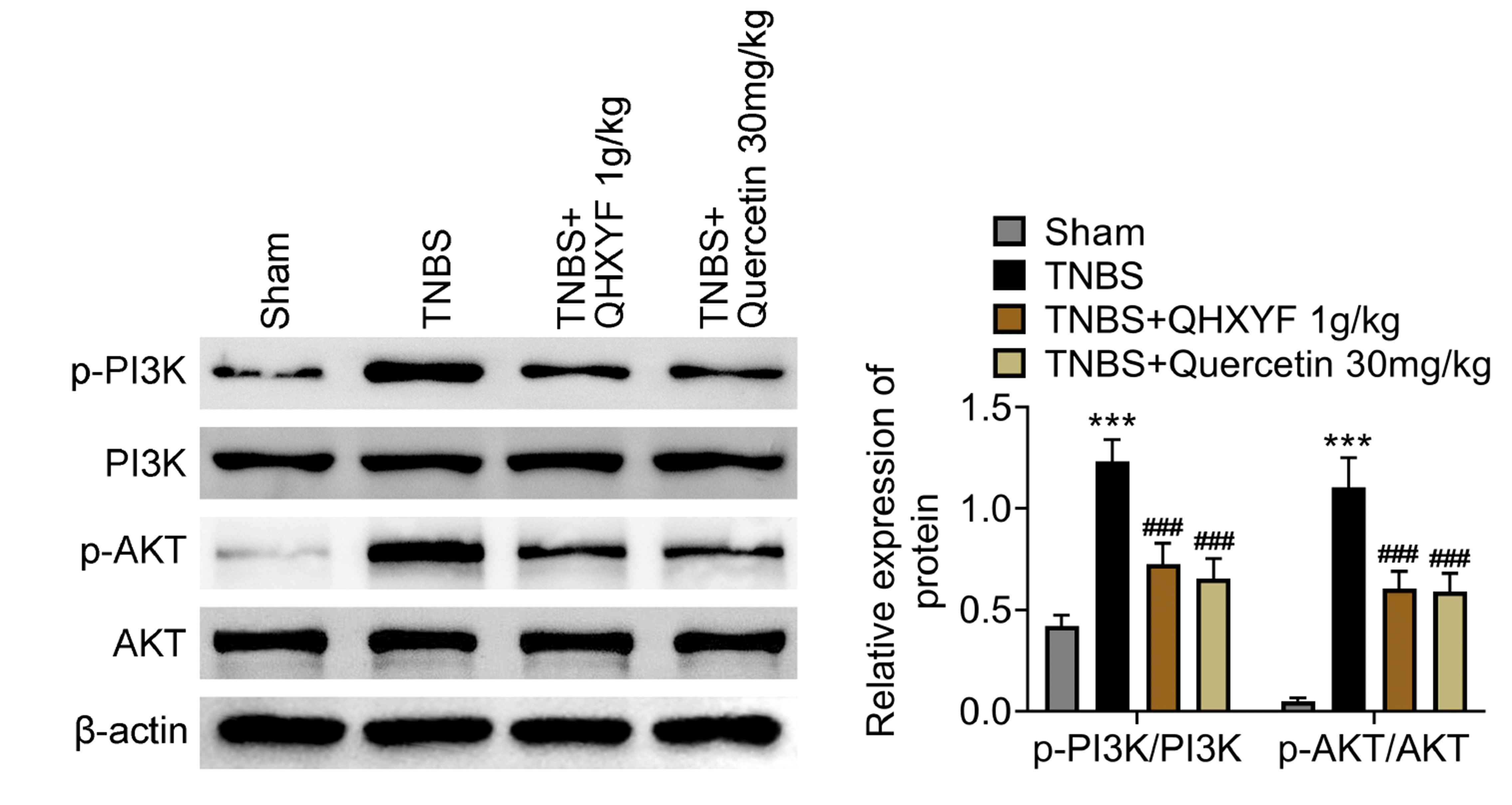
As aforementioned, a total of 166 overlapping targets were identified between QHXYF-related and CD-related disease target data sets and these targets were enriched in the PI3K/AKT pathway. Therefore, we investigated whether QHXYF or quercetin could regulate the PI3K/AKT pathway using Western blot. The results are shown in Fig. 6. We found that TNBS treatment promoted the levels of p-PI3K/PI3K and p-AKT/AKT proteins, while QHXYF or quercetin reduced TNBS-induced increase in these protein levels. In summary, QHXYF or quercetin could inactivate TNBS-induced PI3K/AKT signaling pathway.
CD is a chronic inflammatory illness of the digestive system with an unknown etiology. The development of CD is affected by complex factors such as genetics, immune response, environmental factors, and microbiology [16]. To date, there is no effective treatment available for CD.
TCM has been increasingly recognised as a promising therapeutic approach for CD and colitis-related diseases. However, due to the complex nature of herbal compound therapy with multiple components and targets, understanding the mechanism of TCM therapy can be challenging, especially when the disease pathogenesis is not fully understood. In this context, network pharmacology is a promising approach that can help to elucidate underlying mechanisms and identify important bioactive compounds from TCM formulations [17].
QHXYF is a herbal compound comprises S. baicalensis (Huangqin), C. chinensis (Huanglian), and P. cortex (Huangbai), and has been used to treat UC in clinical practice. However, the exact mechanism of QHXYF in the treatment of CD is yet to be determined. Therefore, we used the TCMSP database to screen for bioactive compounds in QHXYF, and identified a total of 74 bioactive compounds, including quercetin, wogonin, baicalein, and coptisine. Furthermore, we identified 166 targets that were common to QHXYF-related and CD-related disease targets, which could be used for further analysis.
To further confirm whether the 166 overlapping targets enriched in biological activities were associated with CD, we performed GO and KEGG enrichment analyses. The results of the BP enrichment indicated that many of these genes were associated with oxidative stress-related processes, such as “regulation of reactive oxygen species metabolic process”, “cellular response to oxidative stress”, “response to oxidative stress”, among others. Previous studies have shown the digestive system is susceptible to attacks by reactive oxygen species (ROS) [18]. Oxidative stress is a significant contributor to the pathophysiology of gastrointestinal mucosal illnesses, such as colitis, and is caused by shifts in the equilibrium between ROS generation and the capacity to quickly detoxify reactive intermediates [19]. Previous studies have also shown that CD patient sera have high ROS levels and have a pro-oxidative and cytotoxic effect on endothelial cells (ECs) cultured from CD patient sera, resulting in oxidative stress, CD serum-induced oxidative stress and ECs death [20].
In addition, activation of Nrf2 promotes the expression of antioxidant enzymes, leading to the removal of ROS and inhibition of NF-κB activation, ameliorating the clinical symptoms of DSS-induced colitis in mice [21]. The KEGG enrichment results indicated that the 166 overlapping targets were enriched in inflammation-related pathways, such as “IL-17 signaling pathway”, “TNF signaling pathway” and so forth. Complex changes in the inflammatory response, which are defined by changes in the intestinal mucosal barrier's innate immunity and remodeling of the extracellular matrix through increased production of metalloproteins and adhesion molecules, such as MAcCAM-1, contribute to the development of CD [22]. Therefore, inhibition of inflammatory pathways is one of the strategies to treat colitis.
Previous studies have found that inhibiting the IL-17/IL-17R pathway improves Th1 cell chemotaxis and differentiation, suppresses the inflammatory response, improves immune function, and ultimately reduces the severity of CD disease [23]. In clinical practice, anti-TNF therapy has been effective in inducing remission in 60% of patients with CD [24]. Furthermore, therapies for CD that are administered systemically and effectively, such as TNF signaling inhibitors infliximab, adalimumab, and others, have been developed [25].
In addition, KEGG enrichment analysis revealed that most genes were enriched in the “PI3K/AKT signaling pathway” and the etiology of CD was significantly regulated by this pathway. It has been reported that activation of the PI3K/AKT signaling pathway leads to impaired immune tolerance and elevated inflammation while promoting epithelial-mesenchymal transition in intestinal epithelial cells, causing the exacerbation of CD symptoms [26]. Therefore, the results of our enrichment analysis are consistent with the previous studies, which provide a theoretical basis that could be used for future experiments.
Subsequently, Sankey plots and topological analysis by NetworkAnalyzer tools in Cytoscape 3.9.1 showed that quercetin was associated with most of the key bioactive compounds. Quercetin, a polyhydroxyflavonoid, is known for its potent pharmacological effects. According to previous studies, quercetin protects the kidneys by decreasing oxidative stress, enhancing cellular antioxidant capacity, regulating inflammatory factors, and delaying fibrosis by preventing the transformation of renal tubular epithelial cells into myofibroblasts [27]. Quercetin was identified as the most important component of Huai Hua San through network pharmacology, and it was found to inhibit LPS-induced inflammatory factors and the PI3K/AKT signaling pathway in macrophages [28]. Moreover, quercetin was shown to reduce DSS-induced colitis in mice by enhancing intestinal integrity and hepatic antioxidant capacity [29]. These findings collectively suggest that quercetin has anti-inflammatory, anti-oxidative stress, and anti-colitis effects. Consistent with previous studies, this study also demonstrated that quercetin could inhibit TNBS-induced inflammatory response and oxidative stress and alleviate CD symptoms. Furthermore, both molecular docking analysis and western blot showed that quercetin regulated the PI3K/AKT pathway, which is in line with previous reports.
However, the present study has some limitations. Firstly, we only validated one of the predicted enriched pathways experimentally. Secondly, we lacked a significant amount of clinical data. Therefore, future studies should focus on the clinical aspects and explore more potential mechanisms of QHXYF and quercetin.
Overall, this study evaluated the pharmacological mechanism of QHXYF on CD using network pharmacological analysis and molecular docking techniques, followed by experimental validation. We found that QHXYF and its bioactive compound, quercetin, could alleviate TNBS-induced inflammatory response and oxidative stress, and improve CD symptoms in mice by inhibiting the PI3K/AKT pathway.
Supplementary data including one figure can be found with this article online at https://doi.org/10.4196/kjpp.2023.27.4.365.
REFERENCES
1. Roda G, Chien Ng S, Kotze PG, Argollo M, Panaccione R, Spinelli A, Kaser A, Peyrin-Biroulet L, Danese S. 2020; Crohn's disease. Nat Rev Dis Primers. 6:22. Erratum in: Nat Rev Dis Primers. 2020;6:26. Erratum in: Nat Rev Dis Primers. 2020;6:42. Erratum in: Nat Rev Dis Primers. 2020;6:51. DOI: 10.1038/s41572-020-0156-2. PMID: 32242028.

2. Burge K, Gunasekaran A, Eckert J, Chaaban H. 2019; Curcumin and intestinal inflammatory diseases: molecular mechanisms of protection. Int J Mol Sci. 20:1912. DOI: 10.3390/ijms20081912. PMID: 31003422. PMCID: PMC6514688. PMID: c9370cbd52b04bb7a22fc1de731c284e.

3. Ballini A, Santacroce L, Cantore S, Bottalico L, Dipalma G, Topi S, Saini R, De Vito D, Inchingolo F. 2019; Probiotics efficacy on oxidative stress values in inflammatory bowel disease: a randomized double-blinded placebo-controlled pilot study. Endocr Metab Immune Disord Drug Targets. 19:373–381. DOI: 10.2174/1871530319666181221150352. PMID: 30574857.

4. Yu B, Yin YX, Tang YP, Wei KL, Pan ZG, Li KZ, Guo XW, Hu BL. 2021; Diagnostic and predictive value of immune-related genes in Crohn's disease. Front Immunol. 12:643036. DOI: 10.3389/fimmu.2021.643036. PMID: 33936061. PMCID: PMC8085323. PMID: 51f8ee1dadc144dab1d5a60e6a48dd87.

5. Chen M, Ding Y, Tong Z. 2020; Efficacy and safety of Sophora flavescens (Kushen) based traditional Chinese medicine in the treatment of ulcerative colitis: clinical evidence and potential mechanisms. Front Pharmacol. 11:603476. DOI: 10.3389/fphar.2020.603476. PMID: 33362558. PMCID: PMC7758483. PMID: fde00a9930194e1d9ceaa5a2b9a9df08.

6. Xu M, Duan XY, Chen QY, Fan H, Hong ZC, Deng SJ, Nan Z, Wu H, Dong YL, Liu YJ, Zhou CZ. 2019; Effect of compound sophorae decoction on dextran sodium sulfate (DSS)-induced colitis in mice by regulating Th17/Treg cell balance. Biomed Pharmacother. 109:2396–2408. DOI: 10.1016/j.biopha.2018.11.087. PMID: 30551499.

7. Wan F, Wang M, Zhong R, Chen L, Han H, Liu L, Zhao Y, Lv H, Hou F, Yi B, Zhang H. 2022; Supplementation with Chinese medicinal plant extracts from Lonicera hypoglauca and Scutellaria baicalensis mitigates colonic inflammation by regulating oxidative stress and gut microbiota in a colitis mouse model. Front Cell Infect Microbiol. 11:798052. DOI: 10.3389/fcimb.2021.798052. PMID: 35059326. PMCID: PMC8763710. PMID: bc8c567eff69460f94372046c230bd61.

8. Xie Q, Li H, Ma R, Ren M, Li Y, Li J, Chen H, Chen Z, Gong D, Wang J. 2022; Effect of Coptis chinensis franch and Magnolia officinalis on intestinal flora and intestinal barrier in a TNBS-induced ulcerative colitis rats model. Phytomedicine. 97:153927. DOI: 10.1016/j.phymed.2022.153927. PMID: 35030387.

9. Mao YF, Li YQ, Zong L, You XM, Lin FQ, Jiang L. 2010; Methanol extract of Phellodendri cortex alleviates lipopolysaccharide-induced acute airway inflammation in mice. Immunopharmacol Immunotoxicol. 32:110–115. DOI: 10.3109/08923970903193325. PMID: 19811108.

10. Ru J, Li P, Wang J, Zhou W, Li B, Huang C, Li P, Guo Z, Tao W, Yang Y, Xu X, Li Y, Wang Y, Yang L. 2014; TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 6:13. DOI: 10.1186/1758-2946-6-13. PMID: 24735618. PMCID: PMC4001360.

11. Liu J, Zhang L, Wang Z, Chen S, Feng S, He Y, Zhang S. 2022; Network pharmacology-based strategy to identify the pharmacological mechanisms of Pulsatilla decoction against Crohn's disease. Front Pharmacol. 13:844685. DOI: 10.3389/fphar.2022.844685. PMID: 35450039. PMCID: PMC9016333. PMID: 1ef87403ce8f4b2682415fb9acb85ae4.

12. Qi X, Qi C, Kang X, Hu Y, Han W. 2020; Identification of candidate genes and prognostic value analysis in patients with PDL1-positive and PDL1-negative lung adenocarcinoma. PeerJ. 8:e9362. DOI: 10.7717/peerj.9362. PMID: 32607285. PMCID: PMC7315620. PMID: fb375c74400141fda27f5a0ab4220e25.

13. Liu Y, Grimm M, Dai WT, Hou MC, Xiao ZX, Cao Y. 2020; CB-Dock: a web server for cavity detection-guided protein-ligand blind docking. Acta Pharmacol Sin. 41:138–144. DOI: 10.1038/s41401-019-0228-6. PMID: 31263275. PMCID: PMC7471403.

14. Cooper HS, Murthy SN, Shah RS, Sedergran DJ. 1993; Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab Invest. 69:238–249. PMID: 8350599.
15. Lu S, Guo M, Fan Z, Chen Y, Shi X, Gu C, Yang Y. 2019; Elevated TRIP13 drives cell proliferation and drug resistance in bladder cancer. Am J Transl Res. 11:4397–4410. PMID: 31396344. PMCID: PMC6684882.
16. Shen M, Zhang B, Wang M, Meng L, Lv B. 2020; Mica can alleviate TNBS-induced colitis in mice by reducing angiotensin II and IL-17A and increasing angiotensin-converting enzyme 2, angiotensin 1-7, and IL-10. Mediators Inflamm. 2020:3070345. DOI: 10.1155/2020/3070345. PMID: 33100902. PMCID: PMC7569463. PMID: 9cbf498cc8ea485aa69914804ce72a27.

17. Zuo H, Zhang Q, Su S, Chen Q, Yang F, Hu Y. 2018; A network pharmacology-based approach to analyse potential targets of traditional herbal formulas: an example of Yu Ping Feng decoction. Sci Rep. 8:11418. DOI: 10.1038/s41598-018-29764-1. PMID: 30061691. PMCID: PMC6065326.

18. Luceri C, Bigagli E, Agostiniani S, Giudici F, Zambonin D, Scaringi S, Ficari F, Lodovici M, Malentacchi C. 2019; Analysis of oxidative stress-related markers in Crohn's disease patients at surgery and correlations with clinical findings. Antioxidants (Basel). 8:378. DOI: 10.3390/antiox8090378. PMID: 31489956. PMCID: PMC6771139. PMID: 1c479193c06041068ce1facb08f4e2bc.

19. Bhattacharyya A, Chattopadhyay R, Mitra S, Crowe SE. 2014; Oxidative stress: an essential factor in the pathogenesis of gastrointestinal mucosal diseases. Physiol Rev. 94:329–354. DOI: 10.1152/physrev.00040.2012. PMID: 24692350. PMCID: PMC4044300.

20. Ramli I, Posadino AM, Zerizer S, Spissu Y, Barberis A, Djeghim H, Azara E, Bensouici C, Kabouche Z, Rebbas K, D'hallewin G, Sechi LA, Pintus G. 2023; Low concentrations of Ambrosia maritima L. phenolic extract protect endothelial cells from oxidative cell death induced by H2O2 and sera from Crohn's disease patients. J Ethnopharmacol. 300:115722. DOI: 10.1016/j.jep.2022.115722. PMID: 36115603.
21. Deng Z, Cui C, Wang Y, Ni J, Zheng L, Wei HK, Peng J. 2020; FSGHF3 and peptides, prepared from fish skin gelatin, exert a protective effect on DSS-induced colitis via the Nrf2 pathway. Food Funct. 11:414–423. DOI: 10.1039/C9FO02165E. PMID: 31825438.

22. Petagna L, Antonelli A, Ganini C, Bellato V, Campanelli M, Divizia A, Efrati C, Franceschilli M, Guida AM, Ingallinella S, Montagnese F, Sensi B, Siragusa L, Sica GS. 2020; Pathophysiology of Crohn's disease inflammation and recurrence. Biol Direct. 15:23. DOI: 10.1186/s13062-020-00280-5. PMID: 33160400. PMCID: PMC7648997. PMID: 3219b42f20bc4beabc767a488a230c1a.

23. Xia Y, Chen H, Xiao H, Yang J, Li Z, Wang Y, Yang T, Wang B. 2019; Immune regulation mechanism of vitamin D level and IL-17/IL-17R pathway in Crohn's disease. Exp Ther Med. 17:3423–3428. DOI: 10.3892/etm.2019.7389. PMID: 30988721. PMCID: PMC6447769.
24. van Haaften WT, Mortensen JH, Dige AK, Grønbæk H, Hvas CL, Bay-Jensen AC, Karsdal MA, Olinga P, Manon-Jensen T, Dijkstra G. 2020; Serological biomarkers of tissue turnover identify responders to anti-TNF therapy in Crohn's disease: a pilot study. Clin Transl Gastroenterol. 11:e00217. DOI: 10.14309/ctg.0000000000000217. PMID: 33094957. PMCID: PMC7494148.

25. Xu L, Zhang J, Wang Y, Zhang Z, Wang F, Tang X. 2021; Uncovering the mechanism of Ge-Gen-Qin-Lian decoction for treating ulcerative colitis based on network pharmacology and molecular docking verification. Biosci Rep. 41:BSR20203565. DOI: 10.1042/BSR20203565. PMID: 33409535. PMCID: PMC7876598.

26. Wang Z, Zhou H, Cheng F, Zhang Z, Long S. 2022; miR-21 negatively regulates the PTEN-PI3K-Akt-mTOR signaling pathway in Crohn's disease by altering immune tolerance and epithelial-mesenchymal transition. Discov Med. 34:45–58. PMID: 36494326.
27. Tu H, Ma D, Luo Y, Tang S, Li Y, Chen G, Wang L, Hou Z, Shen C, Lu H, Zhuang X, Zhang L. 2021; Quercetin alleviates chronic renal failure by targeting the PI3k/Akt pathway. Bioengineered. 12:6538–6558. DOI: 10.1080/21655979.2021.1973877. PMID: 34528858. PMCID: PMC8806539. PMID: f3957c2a5c3341c2af0852bdd63b7f71.

28. Liu J, Liu J, Tong X, Peng W, Wei S, Sun T, Wang Y, Zhang B, Li W. 2021; Network pharmacology prediction and molecular docking-based strategy to discover the potential pharmacological mechanism of Huai Hua San against ulcerative colitis. Drug Des Devel Ther. 15:3255–3276. DOI: 10.2147/DDDT.S319786. PMID: 34349502. PMCID: PMC8326529.

29. Dong Y, Lei J, Zhang B. 2020; Dietary quercetin alleviated DSS-induced colitis in mice through several possible pathways by transcriptome analysis. Curr Pharm Biotechnol. 21:1666–1673. DOI: 10.2174/1389201021666200711152726. PMID: 32651963.

Fig. 1
Prediction of QHXYF drug targets and CD disease overlapping targets.
(A) Venn diagram showing the intersections of CD-related genes identified from five databases (OMIM, GeneCards, TTD, PharmGkb, and DrugBank). (B) Venn diagram showing the intersection of QHXYF-related and CD-related disease targets. (C) The compound-disease target network depicts the herbal (yellow squares), potential targets (green diamonds), and drug compounds (arrows). (D) NetworkAnalyzer tools in Cytoscape 3.9.1 identified the top 4 core bioactive compounds, and their information is presented. QHXYF, Qinghua Xiaoyong Formula; CD, Crohn’s disease.
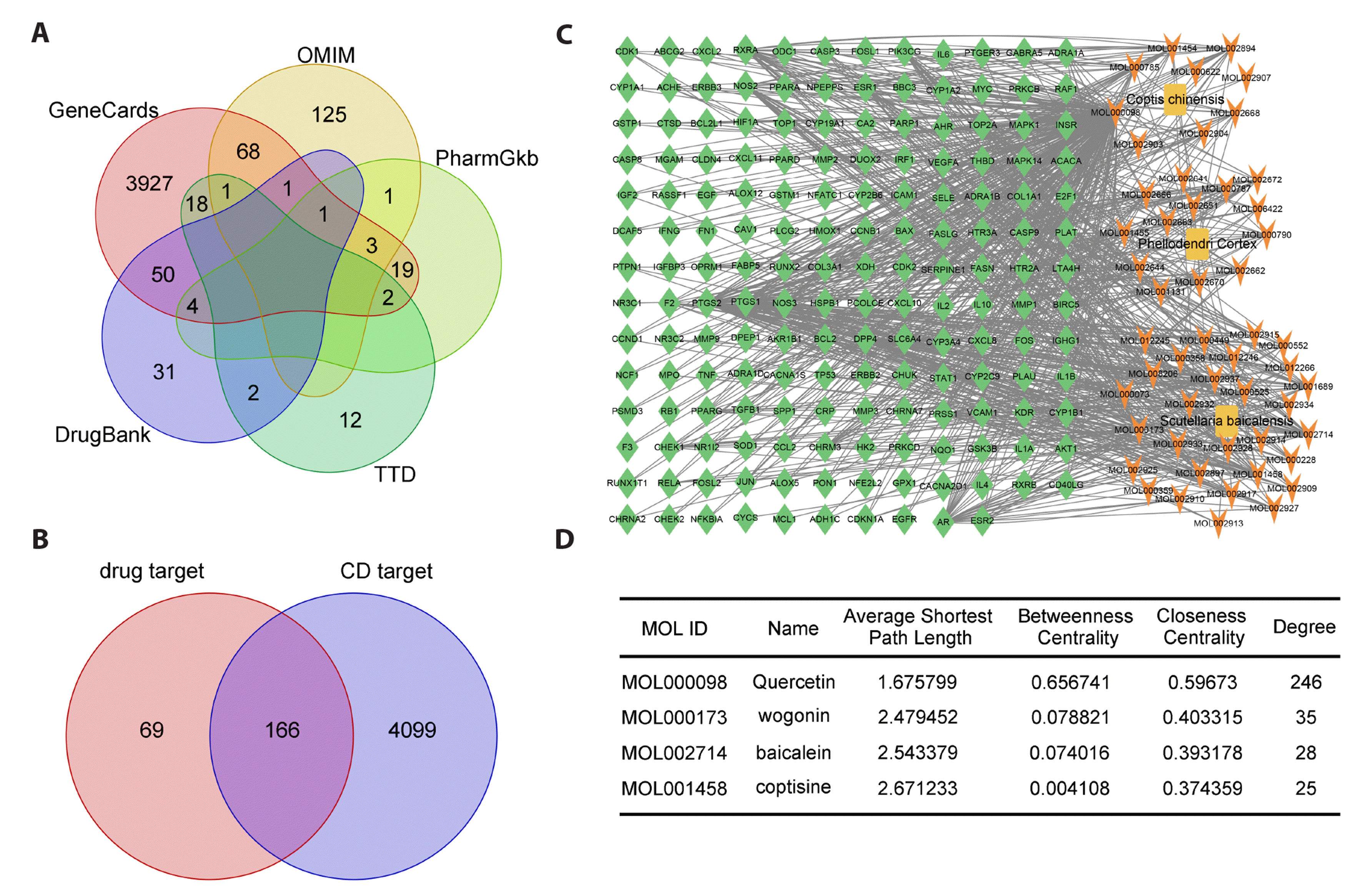
Fig. 2
Functional enrichment analysis and PPI network.
(A) GO analysis of the 166 overlapping targets. (B) KEGG analysis of the 166 overlapping targets. (C) PPI network analysis identifies the key gene targets. Node size indicates the degree of values in descending order, and the darker the color, the more significant the target is. PPI, protein–protein interaction; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
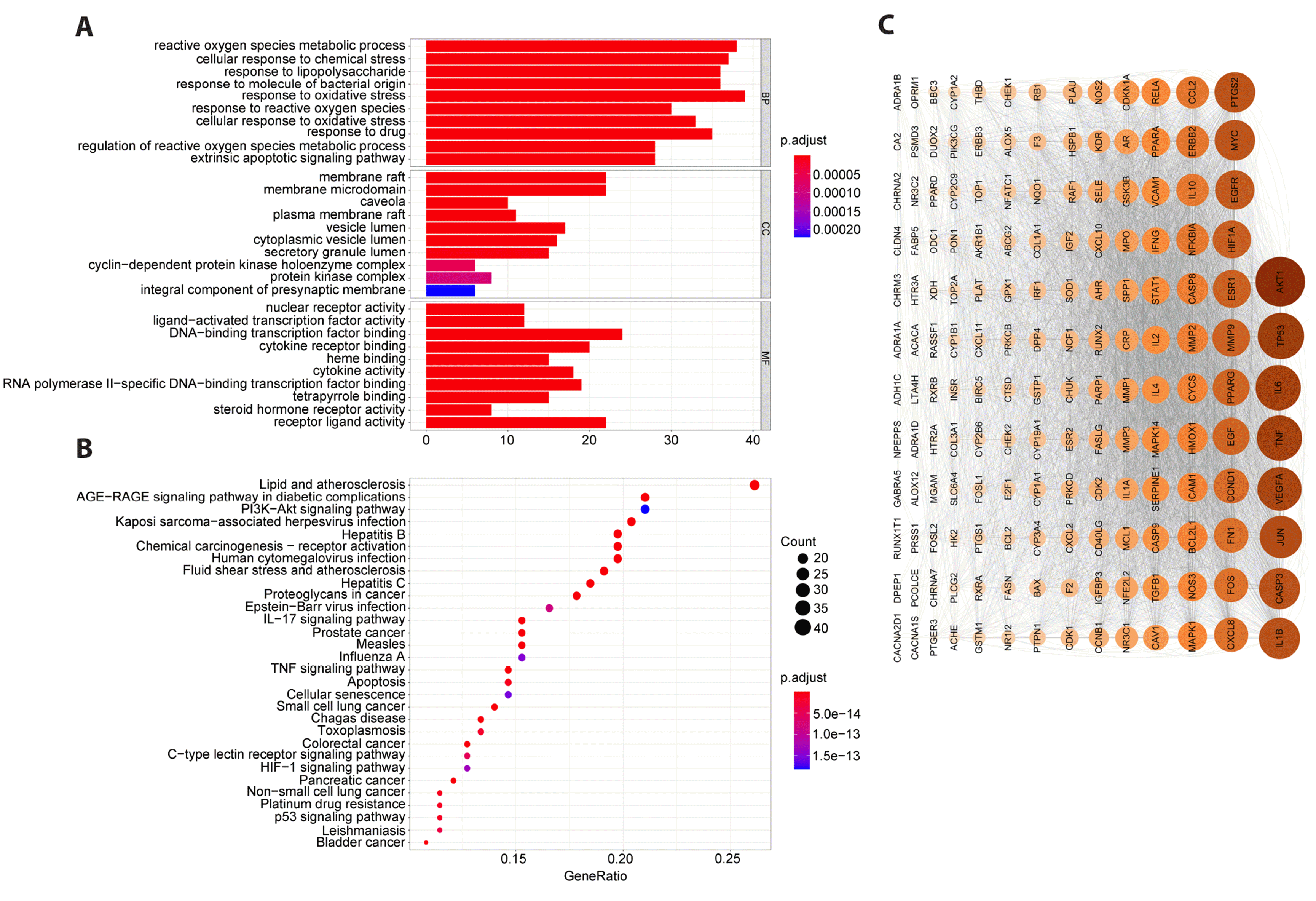
Fig. 3
Molecular docking study of hub genes and key bioactive compounds.
(A) The 10 hub genes identified from the PPI network. The darker the color, the more significant the gene is. (B) Specific rankings and scores of the 10 hub genes. (C) The Sankey diagram reveals the relationship between herbs, compounds, and the top 5 hub targets. The left blocks represent the herb, the middle blocks represent the compounds, and the right blocks represent the hub targets. (D) The results of cavity-detection guided blind docking. (E, F) Quercetin binds to AKT1, TP53, TNF, IL6, and VEGFA. PPI, protein–protein interaction.
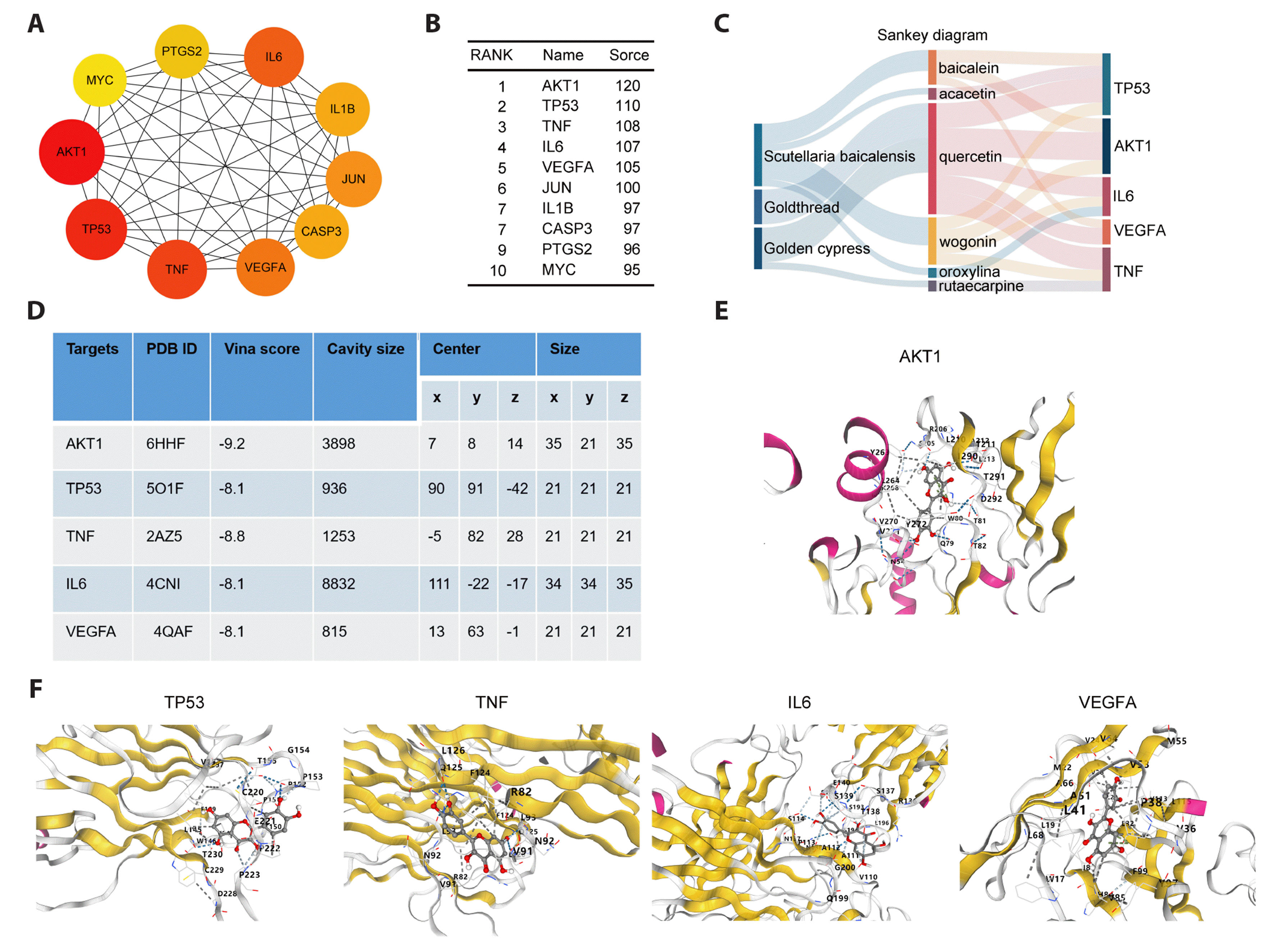
Fig. 4
QHXYF or quercetin alleviates the symptoms of TNBS-induced colitis.
(A) Changes in body weight of TNBS-induced mice treated by QHXYF or quercetin. (B) Disease activity index (DAI) of TNBS-induced mice treated by QHXYF or quercetin. (C) Colon length of TNBS-induced mice treated by QHXYF or quercetin. (D) Pathological changes in colons evaluated using H&E staining. Scale bar = 200 µm. QHXYF, Qinghua Xiaoyong Formula; TNBS, 2,4,6-trinitrobenzenesulfonic acid. ***p < 0.001 compared to the sham group; ###p < 0.001 compared to the TNBS-induced group.

Fig. 5
QHXYF or quercetin inhibits inflammation and oxidative stress in TNBS-induced mouse model.
(A) Assessment of inflammatory factor levels (TNF-α, IL-6, and IL-1β) in serum using ELISA. (B) Assessment of oxidative stress factor levels (MDA, SOD, and GSH) in colon tissues using ELISA. QHXYF, Qinghua Xiaoyong Formula; TNBS, 2,4,6-trinitrobenzenesulfonic acid; TNF-α, tumor necrosis factor-α; IL, interleukin; MDA, monochrome adapter; SOD, superoxide dismutase; GSH, glutathione. ***p < 0.001 compared to the sham group; ###p < 0.001 compared to the TNBS-induced group.
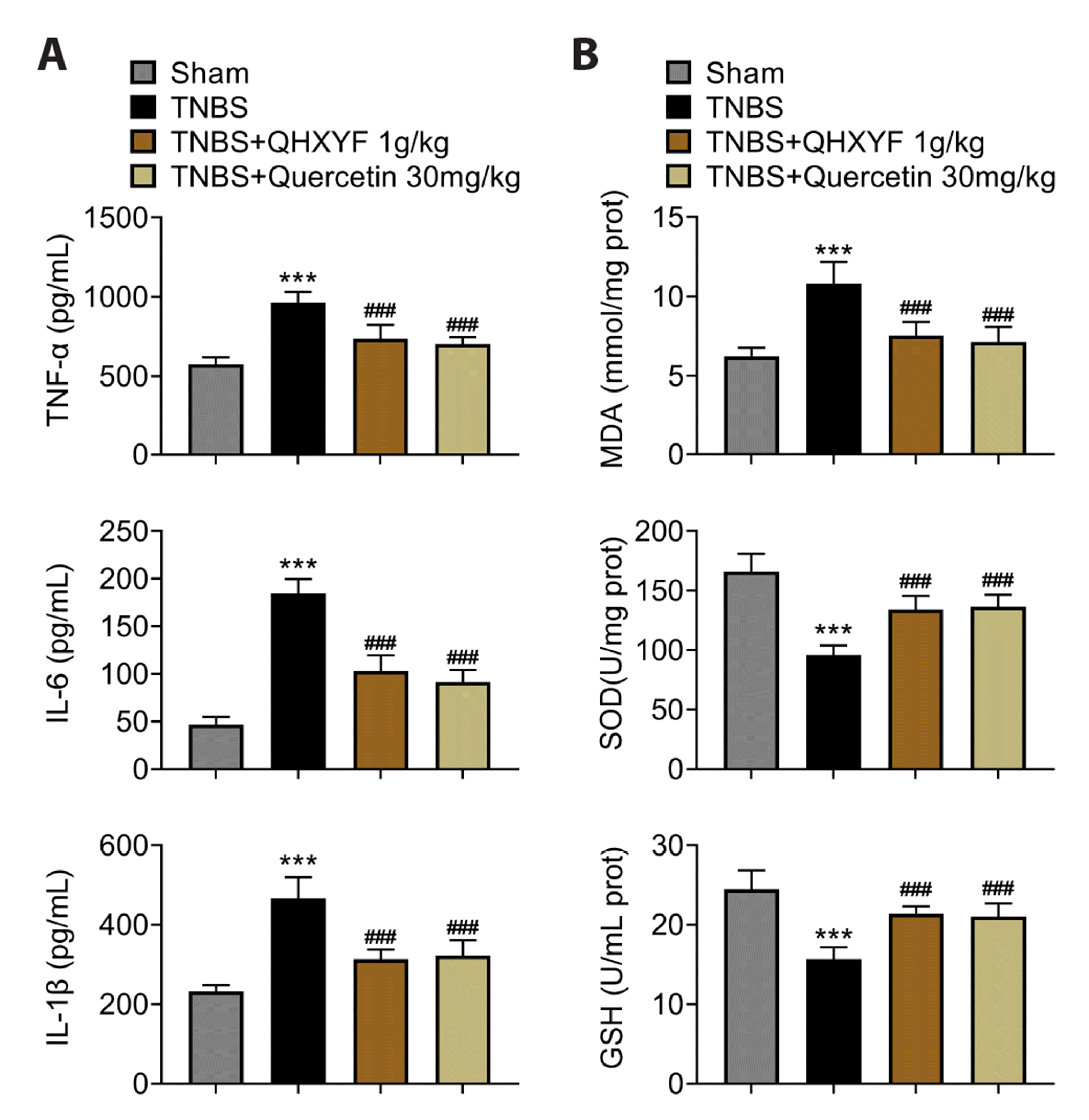
Fig. 6
QHXYF or quercetin regulates the PI3K/AKT signaling pathway.
Assessment of the protein levels of p-PI3K, PI3K, p-AKT, and AKT in TNBS-induced mouse model treated with QHXYF or quercetin using Western blotting. QHXYF, Qinghua Xiaoyong Formula; TNBS, 2,4,6-trinitrobenzenesulfonic acid. ***p < 0.001 compared to the sham group; ###p < 0.001 compared to the TNBS-induced group.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download