Abstract
Cancer initiation and progression are profoundly along with the crosstalk between cancer cells and the surrounding stroma. Accumulating evidence has shown that the therapy targeting the extracellular matrix (ECM) would regress tumor growth and invasion in the most common carcinomas. However, it remains largely unexplored in several rare tumors like odontogenic tumors. Ameloblastoma (AM) is the representative odontogenic epithelial tumor in the jawbone, and it usually infiltrates into adjacent bone marrow and has unlimited growth capacity and a high potential for recurrence. This study aims to investigate the role of collagen-rich ECM during the invasion of AM. Transcriptomic analysis revealed that ECM- and epithelial-to-mesenchymal transition (EMT)-related genes were up-regulated in AM compared to ameloblastoma cell line, AM-1. Tumoroid forming analysis showed that Collagen-rich ECM is indispensable for AM progression, especially for aggressive growth patterns and collective invasion.
Although generally classified as a benign odontogenic tumor, ameloblastoma (AM) presents a high capacity for local infiltration into the adjacent bone marrow with the potential for malignant transformation (1). The extrace-llular matrix (ECM) is a major structural component of the tumor microenvironment (TME) and is highly dynamic around epithelial tumor mass (2). Collagens are the main structural elements of the ECM (3) and promote cancer progression by providing mechanical strength and enhancing cancer cell migration (4). For decades, morpho-pathological analysis of odontogenic tumors has been performed. However, only one study experimentally revealed the physiology of the TME on the relationship between epidermal growth factor and matrix metalloprotease (MMP) secretion in AM (5).
AM-1 is an immortalized cell line generated from plexiform AM (6). Although AM-1 is the most widely used cell line for in vitro studies of AM, most studies have been conducted without realizing the TME of AM lesions because of using the conventional two-dimensional (2D) cell culture method. The possibility of three-dimensional (3D)-organoid (tumoroid) formation using an AM-1 cell line was recently disclosed (7).
In the present study, RNA sequencing (RNA-seq) of AM tissue isolated from patients and of AM-1 cell line was performed, and the transcriptomic profile of ECM- and epithelial-to-mesenchymaltransition (EMT)-related genes were analyzed. In addition, AM-1 tumoroids were cultured in various matrix environments, and the response of AM-1 to ECM changes was confirmed.
This study was approved by the Institutional Review Board (IRB) of the University of Yonsei (No. 2-2018-0050) and followed human subject research guidelines and protocols. Three fresh plexiform ameloblastoma tissues were obtained after surgical procedures following appropriate informed consent from patients (Supplementary file, Table S1) who underwent treatment at the Depart-ment of Oral and Maxillofacial Surgery, Yonsei University Dental Hospital. The diagnoses were made by Two independent board-certified oral and maxillofacial pathologists diagnosed with collected ameloblastoma samples.
AM tissue (n=3) and AM-1 cells (n=1) were collected in TRIzolⓇ Reagent (#15596-026; Invitrogen, CA, USA) and stored at −70℃ until RNA extraction. RNA from each set of sample groups was extracted individually according to the instructions of the manufacturer. RNA concentration was assessed using a NanoDropTM spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) to determine the next step. When the total RNA amount of the sample was lower than 3 μg, the sample collection was repeated. For RNA-seq analysis, we prepared mRNA sequencing libraries as paired-end reads with a length of 100 bases using the TruSeq Stranded mRNA Sample Preparation Kit (Illumina, San Diego, CA, United States) according to the protocols of the manufacturer. The mRNA molecules were purified and fragmented from 2 μg of total RNA. The libraries were sequenced as paired-end reads (2×150 bp) using an Illumina HiSeq2500 sequencer (Illumina, CA, USA).
Low-quality reads were filtered according to the following criteria: reads containing more than 10% of skipped bases (marked as “N” s), reads containing more than 40% of bases whose quality scores were less than 20 and reads with average quality scores of less than 20. The quality scores across all bases were calculated using Sanger/Illumina 1.9 encoding. A quality score of 20 indicated a nucleotide accuracy of 99%. The entire filtering process was performed using in-house scripts.
Differentially expressed gene (DEG) analysis was performed using ‘DESeq2’ (8). To prevent the fold change value of the log scale from being calculated as infinite or non-defined by non-reading (0 value of FPKM) and being excluded as non-significant, all the FPKM values were added 1 E-317, which is less than the smallest value (2.489 E-317) among all gene expression estimations. Genes with fold changes of >2.0 or <0.5, and p-value <0.05, were defined as upregulated and downregulated DEGs, respec-tively.
To characterize the identified genes from DEG analysis, Gene Ontology (GO) analysis was performed using “clu-sterProfiler” package v3.16.1 in R v4.0.3 (9). The GO database classifies genes according to the three categories of biological process (BP), cellular component (CC), and molecular function (MF) and provides information on the function of genes. In addition, GO overrepresentation test was performed with the following parameters: GO annotation database (p-value cutoff: 0.05, q-value cutoff: 0.1). All other options were set to the default values.
The immortalized AM-1 cell line was provided kindly by Prof. Hidemitsu Harada at Iwate Medical University and cultured in keratinocyte serum-free medium (17005 042, Gibco, MA, USA) supplemented with 2.5 μg EGF Human Recombinant (10450-013, Gibco, MA, USA), 25 mg Bovine Pituitary Extract (13028-014, Gibco, MA, USA) and 10,000 U/ml penicillin-streptomycin (15140163, Gibco, MA, USA). The cells were cultured in an incubator at 37℃ in a humidified atmosphere with 5% CO2 and used for the following investigations: tumor formation, histological analysis, and live-cell imaging.
The suspended AM-1 cell was directly dispersed into Matrigel (356237, Corning Life Sciences, Corning, NY, USA) or 4 mg/ml type I Collagen gel (354236, Corning Life Sciences, Corning, NY, USA) at 1×104 cells density per drop. The dish was inverted during the solidification of the Matrigel or collagen gel to prevent the cells from attaching to the culture dish. After solidification for 15 min, the mixture of cells in Matrigel was cultured in an AM-1 tumoroid culture medium. Tumoroid formation was observed under a microscope every 2∼3 d and harvested on day 14.
All quantitative results are expressed as the mean±standard deviation. GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA) was used to analyze all data. One-way analysis of variance with Tukey’s multiple comparison test was performed for multiple group comparisons. Statistical significance was set at p-value <0.05.
The other Materials and Methods were described in detail in the Supplementary files.
Comparison among the whole transcriptomic profile of AM samples obtained from three individual patients (AM #1, 2, and 3) and AM-1 cell was visualized as a heatmap and dendrogram (Fig. 1a). The transcriptomic profile showed a clear separation of samples into two clusters although individual variances were observed among the patient samples. Following DEG analysis of the raw data, GO analysis was performed. The GO terms, including extracellular matrix, collagen, and epithelial-to-mesenchy-mal transition, were significantly downregulated in AM-1 cells (Fig. 1b). Each of the five top-ranked GO term lists related to ECM, collagen, and EMT. Then the expression of genes related to each GO terms was presented as heatmaps (Fig. 1c∼e). The number of genes counted in the top five GO terms was 170 for ECM (excluding duplicates), 217 for collagen, and 60 for EMT (Supplemental datasheet). In ECM-related GO terms, matrix modifying enzymes, including MMPs and KLK5 were found, and the genes encoding ECM proteins (ECM1 and ECM2) were also downregulated in AM-1 compared to AM tissue (Fig. 1c). Among the genes in collagen-related GO terms, downregulation of type I collagen alpha 1 (COL1A1) and COL1A2, which constitute type 1 collagen (Col I), and VIMENTIN (VIM) was found (Fig. 1d). In EMT-related terms, in addition to SNAIL1, which is a direct marker of EMT, TWIST, TGFB, and FGFR, which are known to affect or be affected by EMT, were identified (Fig. 1e).
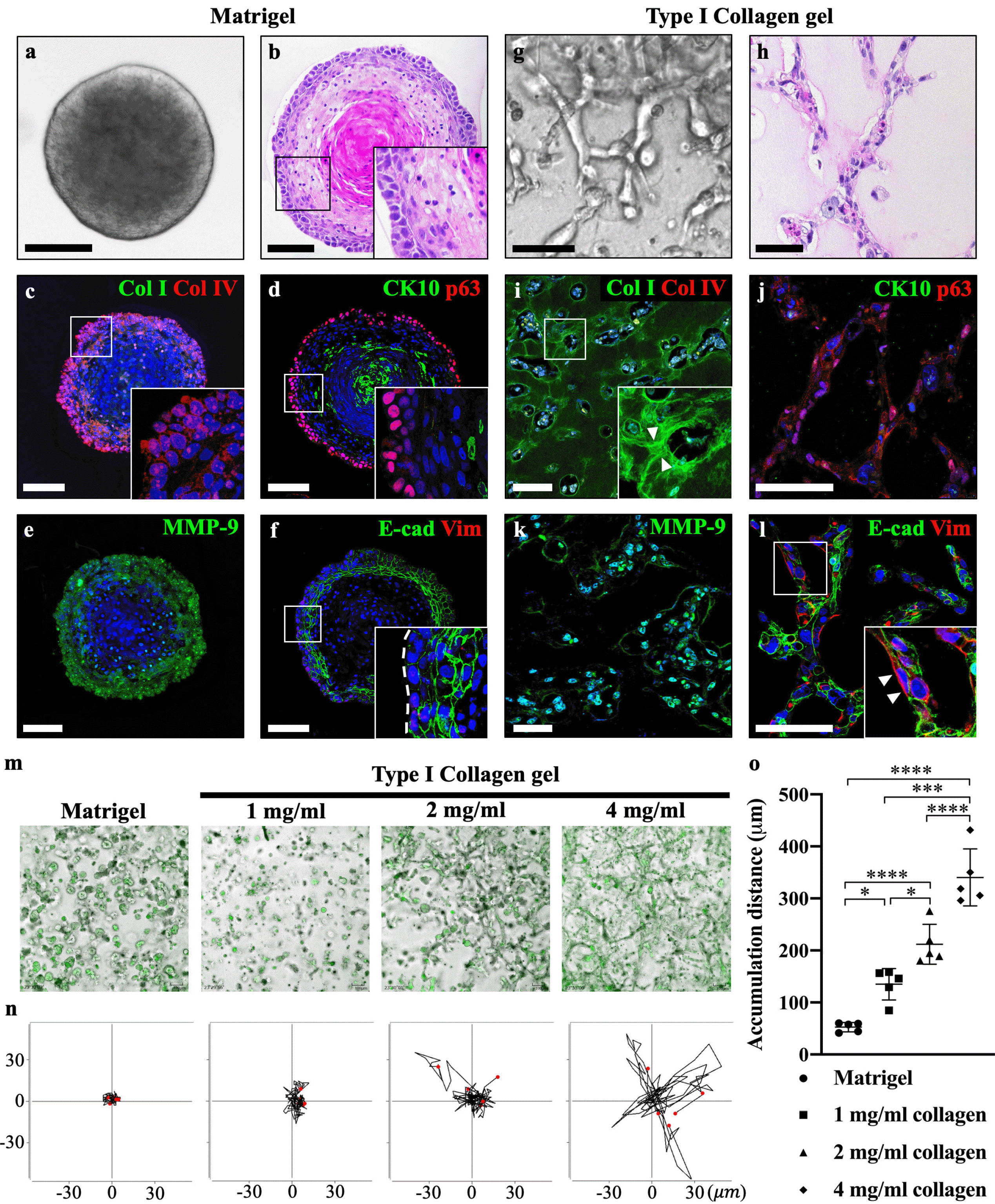
The main difference between Matrigel and collagen gel is the concentration of Col I. Round-shaped tumoroids with well-organized multicellular structures were observed in Matrigel-cultured group (Fig. 2a and 2b). In contrast, collagen gel-cultured tumoroids invaded into the adjacent ECM and generated net-like structures (Fig. 2g and 2h). Interestingly, the expression of type IV collagen (Col IV, a major component of the basement membrane) was intensely expressed in the basal cell layer, whereas Col I was not observed in Matrigel-cultured tumoroids (Fig. 2c). Conversely, intense expression of Col I was observed in collagen gel-cultured tumoroids, while Col IV was negatively expressed (Fig. 2i). And radial-shaped fibrillar collagen was observed at the edge of the invading strands from tumoroids (Fig. 2i, arrowheads). Furthermore, basal cells were detected with p63, and the epidermal differentiation marker cytokeratin 10 (CK10) was observed in the core of the Matrigel-cultured tumoroids (Fig. 2d). In contrast, CK10 was negatively expressed, and moderate expression of p63 was observed in collagen gel-cultured tumoroids (Fig. 2j). The expression of MMP9 was signifi-cantly downregulated in AM-1 cells (Fig. 1c). However, intense expression of MMP-9 was detected in both Matrigel- and collagen gel-cultured tumoroids (Fig. 2e and 2k). 3D culture conditions restored the expression of MMP-9 in AM-1 cells. Despite the expression of MMP-9 was also observed in the Matrigel-cultured tumoroids, a low concentration of Col I as a substrate in Matrigel is insufficient to provide enough mechanical strength for the cancer cell invasion. E-cadherin was intensely expressed in the suprabasal layer and Vimentin was not expressed in Matrigel-cultured tumoroids (Fig. 2f and 2l). Vimentin was expressed in the edge of the invading strands in collagen gel-cultured tumoroids (Fig. 2l, arrowhead).
To confirm the effect of Col I in the invasion of AM-1 cells, varying concentrations (1, 2, and 4 mg/ml) of collagen gel were used in the tumoroid culture system and the mobility of AM-1 cells was analyzed. Compared to the Matrigel, numerous invading strands were observed in the collagen gel-cultured tumoroids, which is similar to the plexiform AM (10) (Fig. 2m). Individual cell migration was tracked within a 2D plane, and the cell trajectory plot indicated that directional cell movement was increased in Col I dose-dependent manner (Fig. 2n). The accumulation distance of cell migration in the collagen gel was significantly higher than that in the Matrigel and increased in a Col I dose-dependent manner (Fig. 2o). These results demonstrate that among the several components of ECM molecules, Col I play a crucial role in promoting the invasive growth pattern of AM-1 tumoroids.
Transcriptomic comparison between AM and AM-1 showed that ECM-, Collagen-, EMT-related GO terms, and related genes were upregulated in the AM compared to the AM-1. Two possible reasons for the results are: 1) the AM tissue sample contains both the epithelial tumor mass and surrounding stromal cells, whereas the AM-1 cell line does not contain fibroblasts that produce collagen fibrils; and 2) the expression of ECM- or EMT-related genes is no longer necessary as the cell were cultured two-dimensionally.
To investigate the possible effect of the culture method on the expression of ECM- and EMT-related genes, AM-1 cell were three-dimensionally cultured in Matrigel or collagen gel, the standard 3D culture method to provide stroma-like environments. Collagen gel-cultured tumoroid showed a more invasive growth pattern compared to the Matrigel-cultured tumoroid. Matrigel provides the connective tissue bed including a basal layer to epithelial cells, whereas collagen gel can mimic tumor-associated collagen architectures (11). Active remodeling of surrounding ECM was observed in the collagen gel-cultured tumoroid, but not in the Matrigel. This phenomenon is similar to the stiffening of the ECM by forming fibrillar collagen when the tumor breaches the basement membrane for invasion (12). In addition, strong CK10 expression was detected in the core of Matrigel-cultured tumoroid. These results demonstrated that basement membrane components provided by Matrigel enhanced the differentiation of AM-1 tumoroids. MMP-9 is a well-known enzyme involved in the proteolytic degradation of ECM during cancer invasion and metastasis (13). The expression of MMP-9 was observed in both Matrigel and collagen gel-cultured tumoroid. However, without the support of the collagen component in the Matrigel, AM-1 tumoroid could not get sufficient mechanical strength for the invasion. Most invasive solid tumors, including AM, display predominantly collective invasion, in which multicellular clusters invade the peritumoral stroma while maintaining cell-cell adhesions (14). In the collagen gel-cultured tumoroid, the collective invasion was observed with the co-localized expression of E-cadherin and vimentin. A decrease in E-cadherin and an increase in vimentin expression in the outermost cells of the tumor mass are typical features of partial EMT and collective invasion (15). To sum up, collagen-rich ECM provides more favorable conditions for the invasion of AM-1 tumoroids than Matrigel. This seems to be due to the differencein stiffness according to the collagen concentration in the ECM.
More importantly, the invasion distance of AM-1 cells was significantly increased in the Col I concentration-de-pendent manner. A higher concentration of Col I may contribute to ECM stiffness, which is one of the most essential extrinsic factors for cancer cell invasion and metastatic progression (16).
In conclusion, AM-1 showed a different transcriptomic pattern from AM tissue, but the expression of genes related to the microenvironment was restored to some extent through 3D tumoroid culture. In addition, the invasion of AM-1 tumoroids into surroundings was increased in a Col I concentration-dependent manner. Collagen-rich ECM is indispensable for AM progression, especially for aggressive growth patterns and collective invasion. Further studies are necessary to confirm whether the different reactivity of AM-1 to collagen is through the response of mechano-transducers due to matrix stiffness and verify this using primary AM cell tumoroids.
Supplementary data including one table and datasheet can be found with this article online at https://doi.org/10. 15283/ijsc22132.
Acknowledgments
This study was supported by the Yonsei University College of Dentistry Fund (6-2019-0014). The skillful assistance for visualizing RNA-seq data of Junsoo Song (NGeneS Inc.) is gratefully acknowledged.
Notes
Ethics Approval
Human surgical specimens: Patients with AM who successfully underwent surgery at Yonsei University Hospital (Seoul, Republic of Korea) between 2019 and 2021 were enrolled under the approval of the Institutional Review Boards of Yonsei University Health System (YUHS-IRB 2-2018-0050).
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
References
1. Effiom OA, Ogundana OM, Akinshipo AO, Akintoye SO. 2018; Ameloblastoma: current etiopathological concepts and ma-nagement. Oral Dis. 24:307–316. DOI: 10.1111/odi.12646. PMID: 28142213. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85014775401&origin=inward.

2. Nallanthighal S, Heiserman JP, Cheon DJ. 2019; The role of the extracellular matrix in cancer stemness. Front Cell Dev Biol. 7:86. DOI: 10.3389/fcell.2019.00086. PMID: 31334229. PMCID: PMC6624409. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85076736643&origin=inward.

3. Frantz C, Stewart KM, Weaver VM. 2010; The extracellular matrix at a glance. J Cell Sci. 123(Pt 24):4195–4200. DOI: 10.1242/jcs.023820. PMID: 21123617. PMCID: PMC2995612. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=78649737455&origin=inward.

4. Xu S, Xu H, Wang W, Li S, Li H, Li T, Zhang W, Yu X, Liu L. 2019; The role of collagen in cancer: from bench to bedside. J Transl Med. 17:309. DOI: 10.1186/s12967-019-2058-1. PMID: 31521169. PMCID: PMC6744664. PMID: b8b51f6937ce41229bd395a7985054b6. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85072165935&origin=inward.

5. da Rosa MR, Falcão AS, Fuzii HT, da Silva Kataoka MS, Ribeiro AL, Boccardo E, de Siqueira AS, Jaeger RG, de Jesus Viana Pinheiro J, de Melo Alves Júnior S. 2014; EGFR signaling downstream of EGF regulates migration, invasion, and MMP secretion of immortalized cell[s derived from human ameloblastoma. Tumour Biol. 35:11107–11120. DOI: 10.1007/s13277-014-2401-3. PMID: 25099616. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84928582417&origin=inward.

6. Harada H, Mitsuyasu T, Nakamura N, Higuchi Y, Toyoshima K, Taniguchi A, Yasumoto S. 1998; Establishment of ameloblastoma cell line, AM-1. J Oral Pathol Med. 27:207–212. DOI: 10.1111/j.1600-0714.1998.tb01943.x. PMID: 9682983. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0032077706&origin=inward.

7. Chang TH, Shanti RM, Liang Y, Zeng J, Shi S, Alawi F, Carrasco L, Zhang Q, Le AD. 2020; LGR5+ epithelial tumor stem-like cells generate a 3D-organoid model for amelobla-stoma. Cell Death Dis. 11:338. DOI: 10.1038/s41419-020-2560-7. PMID: 32382005. PMCID: PMC7206107. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85084403314&origin=inward.
8. Love MI, Huber W, Anders S. 2014; Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. DOI: 10.1186/s13059-014-0550-8. PMID: 25516281. PMCID: PMC4302049. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84924629414&origin=inward.

9. Yu G, Wang LG, Han Y, He QY. 2012; clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16:284–287. DOI: 10.1089/omi.2011.0118. PMID: 22455463. PMCID: PMC3339379. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84860718683&origin=inward.

10. Modolo F, Martins MT, Loducca SV, de Araújo VC. 2004; Expression of integrin subunits alpha2, alpha3, alpha5, alphav, beta1, beta3 and beta4 in different histological types of ameloblastoma compared with dental germ, dental lamina and adult lining epithelium. Oral Dis. 10:277–282. DOI: 10.1111/j.1601-0825.2004.01028.x. PMID: 15315644. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=4644327945&origin=inward.

11. Gong X, Kulwatno J, Mills KL. 2020; Rapid fabrication of collagen bundles mimicking tumor-associated collagen architec-tures. Acta Biomater. 108:128–141. DOI: 10.1016/j.actbio.2020.03.019. PMID: 32194262. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85082822631&origin=inward.

12. Chang TT, Thakar D, Weaver VM. 2017; Force-dependent breaching of the basement membrane. Matrix Biol. 57-58:178–189. DOI: 10.1016/j.matbio.2016.12.005. PMID: 28025167. PMCID: PMC5328923. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85008350831&origin=inward.

13. Xu D, McKee CM, Cao Y, Ding Y, Kessler BM, Muschel RJ. 2010; Matrix metalloproteinase-9 regulates tumor cell invasion through cleavage of protease nexin-1. Cancer Res. 70:6988–6998. DOI: 10.1158/0008-5472.CAN-10-0242. PMID: 20736374. PMCID: PMC3272441. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=77956291596&origin=inward.

14. Friedl P, Locker J, Sahai E, Segall JE. 2012; Classifying collective cancer cell invasion. Nat Cell Biol. 14:777–783. DOI: 10.1038/ncb2548. PMID: 22854810. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84864886120&origin=inward.

15. Vilchez Mercedes SA, Bocci F, Levine H, Onuchic JN, Jolly MK, Wong PK. 2021; Decoding leader cells in collective cancer invasion. Nat Rev Cancer. 21:592–604. DOI: 10.1038/s41568-021-00376-8. PMID: 34239104. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85110033085&origin=inward.

16. Seewaldt V. 2014; ECM stiffness paves the way for tumor cells. Nat Med. 20:332–333. Erratum in: Nat Med 2014;20: 1217. DOI: 10.1038/nm.3523. PMID: 24710372. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84898037346&origin=inward.

Fig. 1
Transcriptomic comparison between ameloblastoma and AM-1 cell line. (a) Heatmap of whole transcriptome showing a significant difference in the transcriptome between three AM samples (AM#1, #2, #3) and AM-1 cell line. (b) Volcano plot of GO term of significantly down-regulated DEGs. The top five GO terms related to ECM (red), collagen (blue), and EMT (green) are indicated in different colors, and the top five GO IDs are selectively labeled. Cutoff values for DEG level in GO analysis were adjusted p-value <0.05 and |fold change| >2.0. (c∼e) Top five GO term lists and heatmaps of genes consisting of top three GO terms related to ECM, collagen, and EMT. In heatmaps, major genes of the tumor microenvironment are labelled.
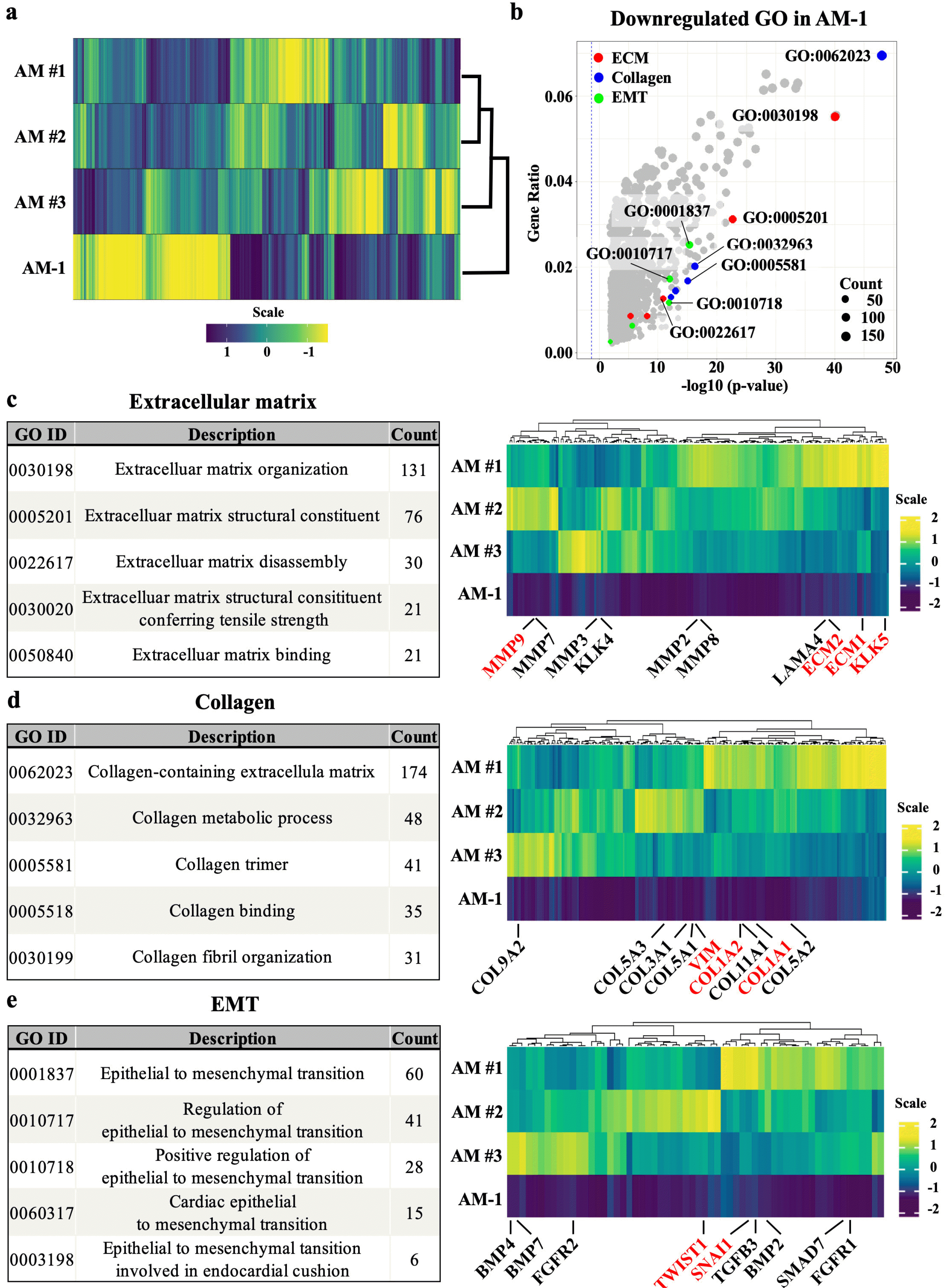
Fig. 2
Type I collagen plays a crucial role in AM-1 tumoroid progression. (a) Brightfield image showing that round-shaped AM-1 tumoroids were observed in the Matrigel culture group. (b) Hematoxylin and eosin staining of a well-organized tumoroid structure, comprised of a heterogeneous cell population. (c) Col I was negatively expressed, and intense expression of Col IV was observed surrounding the edge of Matrigel-cultured tumoroids. (d) The terminal differentiation marker CK10 was strongly expressed in the tumoroid core, and p63 was intensely expressed in the basal cell layer. (e) MMP-9 showed a moderate expression in the basal cell layer. (f) E-cadherin was intensely expressed in the suprabasal layer and moderately expressed in the basal cell layer (indicated by a white dotted line). Vimentin was negatively expressed in the Matrigel culture group. (g) Net-like structures were observed in collagen gel-cultured (4 mg/ml) AM-1 tumoroids. (h) Hematoxylin and eosin staining of the invading strands from collagen gel-cultured tumoroids. (i) Immunohistochemistry staining of Col I and Col IV. Arrowheads indicate the rearranged fibrillar collagen fiber by adjacent AM-1 tumoroids, and Col IV was negatively expressed. (j) CK10 showed negative expression, and p63 showed a moderate expression. (k) Intense MMP-9 expression was observed in the majority of collagen-cultured AM-1 tumoroids. (l) E-cadherin showed a moderate expression, and vimentin was intensely expressed in the leading region of the invading tumor strand as indicated by an arrowhead. Nuclei were counterstained with TO-PRO-3 (TP-3). (m) Representative images of AM-1 cell cultured in Matrigel or varying concentrations of type I collagen gel (1, 2, and 4 mg/ml) obtained from 10 h of live-cell imaging. AM-1 cells were indicated with CMFDA dye (green). (n) Trajectory plots depicted the movement path of individual AM-1 cells (n=5). Through transformation, all trajectories started at the origin (x=0, y=0) in order to visualize the movements of all cells relative to each other. (o) Quantitative analysis of accumulation distance (μm) of Matrigel or collagen gel-cultured AM-1 cell. Scale bars: 100 μm. *p<0.05, ***p<0.001, and ****p<0.0001.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download