INTRODUCTION
Based on the haemagglutinin (H) protein, canine distemper virus (CDV) of the genus Morbillivirus has been classified into several genotypes: America-1 and -2, Asia-1 and -2, Europe, Europe wildlife, Arctic-like, and Africa (1). Genetic lineages are distinguished on the basis of strains showing amino acid divergence in the H protein of less than 4% (2). The H protein attaches to cellular receptors, such as signaling lymphocyte activation molecule (SLAM), CD46, and Nectin-4, thereby playing a role in cell tropism and adaptability (3).
CDV contains a single-stranded negative RNA genome that encodes six structural proteins: nucleocapsid protein (N), phosphoprotein, matrix protein, fusion protein (F), H protein and polymerase large protein (L) (4). The F and H proteins facilitate viral entry into host cells. In particular, as the most conserved morbillivirus (together with the H gene sequence), the F gene sequence has been used to determine genetic associations among different morbillivirus species (5).
Similar to other countries, in South Korea, CDV-related diseases were increasing in prevalence among dog and raccoon dog populations (6). Thus, CDV vaccine strains
such as Lederle and Rockborn were developed in America in the 1950s and were released to prevent CDV infections in South Korea beginning in the 1980s (7). These CDV vaccine strains have shown efficacy in reducing CDV infection rates among Korean dog populations. Due to the development of molecular and biological technologies, gene analysis of CDV-infected dogs has been possible since 2000. Several studies have reported that the Asia-1 and -2 CDV genotypes continue to circulate among populations of raccoon dogs and dogs in South Korea (6, 8). In addition, the wild Korean CD1901 strain, belonging to the Asia-1 genotype and isolated from a naturally infected dog, is expected to be used as a vaccine strain in the future (9).
A method is required to differentiate the CD1901 strain from existing vaccine strains, such as Lederle and Rockborn belonging to the America-1 genotype because the vaccine strain may be shed in vaccinated dogs. Five methods of CDV diagnosis are widely used: rapid immunodiagnostic tests (RIDTs), sandwich enzyme-linked immunosorbent assay, virus isolation, molecular techniques (reverse-transcription polymerase chain reaction [RT-PCR], and real-time RT-PCR), reverse-transcription loop-mediated isothermal amplification, and histological identification (10, 11, 12). However, although these methods can detect the presence of CDV antigens and genes in samples, they cannot differentiate among the CDV genotypes and strains. Therefore, a method for differentiating the CD1901 strain from the Lederle CDV vaccine strain without the need for nucleotide sequence analyses is required. In this study, we established a multiplex RT-PCR method for one-step detection of CDV based on mutation of the CDV F glycoprotein.
MATERIALS AND METHODS
Viruses and cells
CD1901 was isolated from a naturally infected dog in South Korea in 2019 (9). The attenuated CDV vaccine strain, Lederle, introduced to South Korea from the US in the 1980s, has been used to prevent canine distemper. The CD1901 and Lederle strains were propagated in Vero/dSLAM cells expressing the dog SLAM gene in Dulbecco’s modified Eagle’s medium containing two antibiotics, an antifungal agent, and 10% heat-inactivated fetal bovine serum (Gibco BRL, Grand Island, NY, USA). The CD1901 and Lederle strains were used as positive controls for multiplex RT-PCR, and the analytical sensitivity and analytical specificity of the primers were determined. Four commercial distemper/adeno/parvo/parainfluenza (DAPP) vaccines containing CDV, canine adenovirus type 1 (CAV-1) or canine adenovirus type 2 (CAV-2), canine parvovirus (CPV), and canine parainfluenza virus (CPIV) manufactured by Korean biological companies were used for multiplex RT-PCR.
Identification and titration
The CD1901 and Lederle strains were inoculated into Vero/dSLAM cells grown in a 25 cm2 flask, and the cytopathic effects (CPEs) of the two strains were compared. CPEs were observed daily under a microscope; at 5 days post-inoculation, each flask was frozen and thawed three times. The solutions then were subjected to viral titration. The titers of the two viruses were determined according to a previously reported method (9).
Design of primer sets
Primer sets were designed based on the N and F gene sequences of the CD1901 and Lederle strains (GenBank accession no. EF418783). For the forward and reverse primers, positions CD1901-F/-R and Lederle-F-/R were selected based on the nucleotide sequence of the F gene, while the position of CDV complex-R corresponded to the nucleotide sequence of two CDV N gene. The primer sets for CDV complex-F/-R, CD1901-F/-R, and Lederle-F/-R were expected to generate 224-, 326- and 428-bp products, respectively. The sequences and targeted nucleotide positions of the primers are shown in Table 1 and Figs. 1 and 2.
Table 1.
Details of the three multiplex RT-PCR primer sets designed for differentiation between the CD1901 and Lederle CDV strains
| Name of primer | Oligonucleotide sequence (5’-3’) | Expected size (bp) | Target gene | Remarks |
|---|---|---|---|---|
| CDV complex-F | GTTCAAGAGGACTCGGGACCAACC | 224 | N | CD1901 and Lederle |
| CDV complex-R | CAGGGGATTCCACGAACAAGG | |||
| CD1901-F | TAATGATATGATTGCAATTTTGGAGAG | 326 | F | CD1901 |
| CD1901-R | CGCCCCTAATGCATTGTTGTAGAATC | |||
| Lederle-F | GGAGGCAGGCAATACCCTGAT | 428 | F | Lederle |
| Lederle-R | CTTGAAGAAAATTGGGCGGGGTTA |
Fig. 1
Amplification sites of the canine distemper virus (CDV) gene for multiplex reverse-transcription polymerase chain reaction (RT-PCR). Primer sets for discriminating the CD1901 and Lederle strains were designed based on the nucleocapsid (N) and fusion (F) genes.
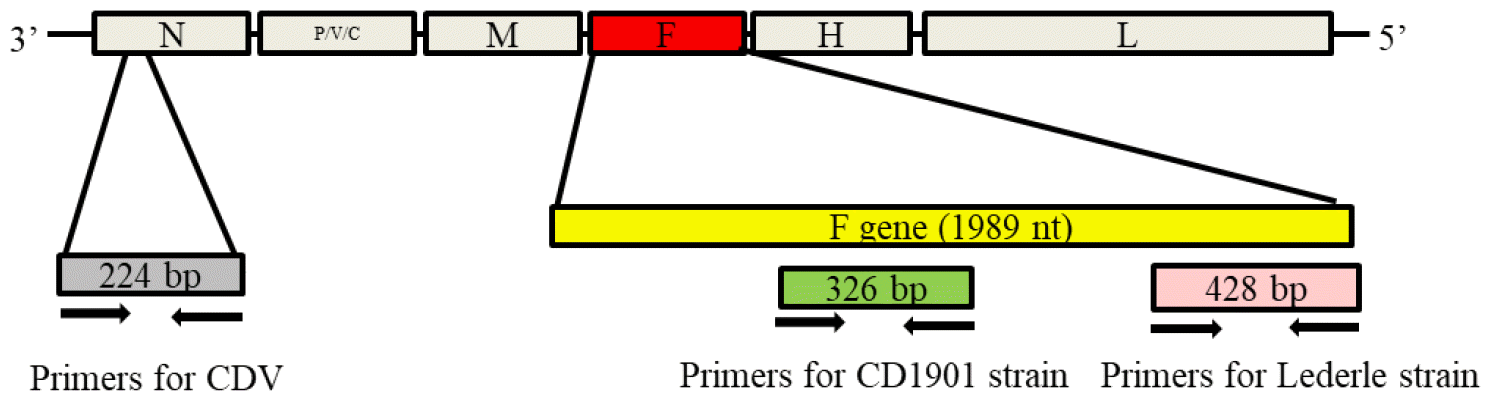
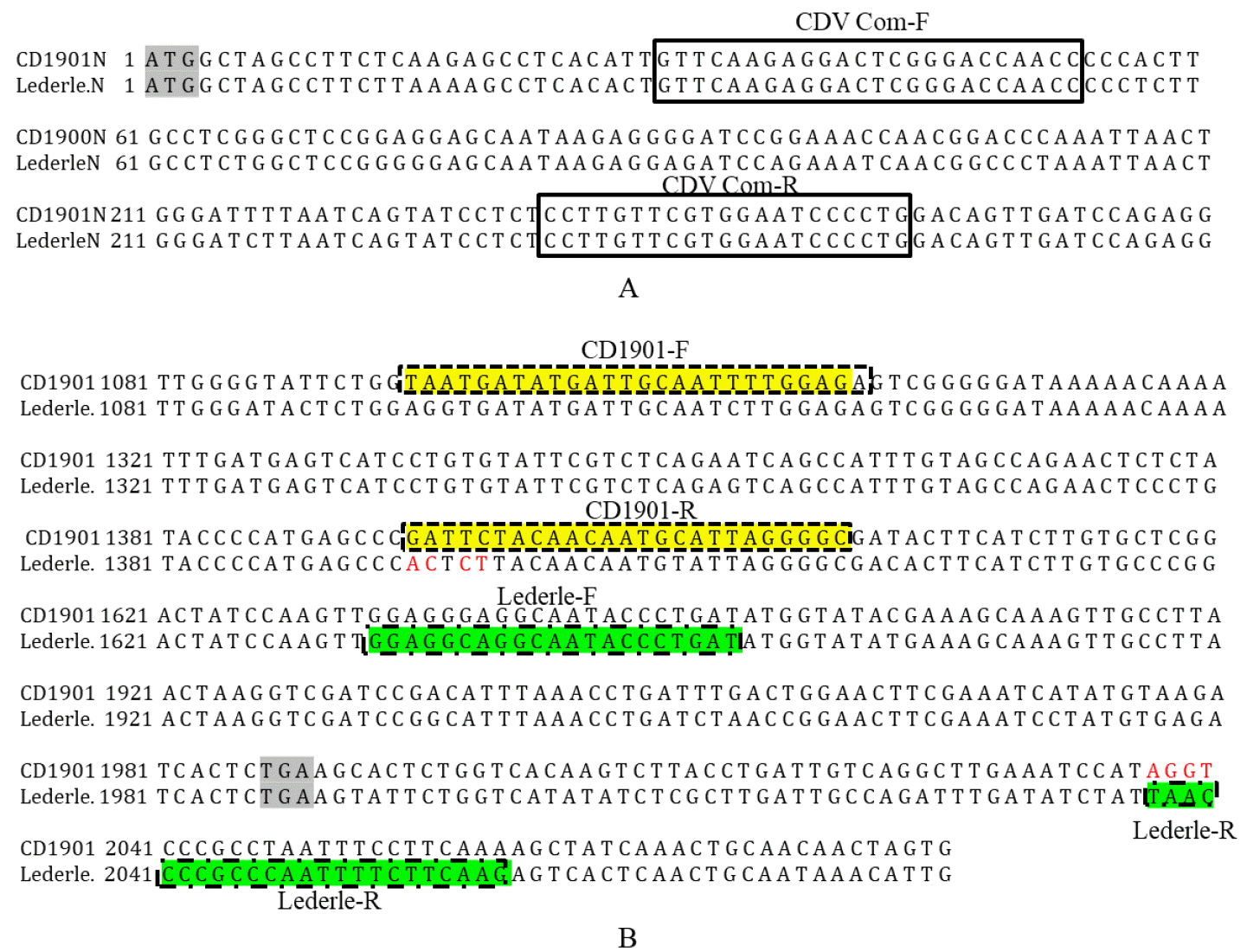
Fig. 2
Canine distemper virus (CDV) fusion protein (F) sites targeted by the multiplex reverse-transcription polymerase chain reaction (RT-PCR) primers. Four of the first five nucleotides (GATTC) of the CD1901-R primer were not identical to those (ACTCT) of the Lederle strain. The first four nucleotides (TAAC) of the Lederle-R primer were not identical to those (AGGT) of the CD1901 strain. ATG marks the beginning of the N gene and TGA means the end of the open reading frame of the F gene.
RNA/DNA extraction and sensitivity of multiplex RT-PCR
Total viral RNA was extracted from the CDV, CPIV, and rabies RNA viruses using an RNA extraction kit (Bioneer, Daejeon, Korea), while genomic DNA was extracted from the CPV, CAV-1, and CAV-2 DNA viruses using genomic DNA extraction kit (Bioneer) in accordance with the manufacturer’s instructions. The extracted RNA/DNA was eluted in 50 µl RNase- and DNase-free water and subjected to RT-PCR. Multiplex RT-PCR was performed using the One Step RT-PCR kit (Qiagen, Hilden, Germany). Each RT-PCR mixture contained 2 µl denatured RNA, 0.5 µl each primer (50 pmol), 5 µl 5× buffer (12.5 mM MgCl2), 2 µl dNTP mix, 1 µl enzyme mix (reverse transcriptase and Taq polymerase), and 13 µl distilled water. The RT-PCR protocol consisted of cDNA synthesis at 50°C for 30 min, followed by 35 cycles of 95°C for 45 s, 55°C for 45 s, and 72°C for 45 s, with a final extension at 72°C for 5 min. The PCR products were electrophoresed on a 2.0% agarose gel for 30 min and visualized using Redsafe™ nucleic acid staining solution (iNtRON Biotechnology, Seoul, Korea).
Specificity and application
Multiplex RT-PCR was performed to detect CDV strains (CD1901, Lederle, and commercial vaccine strains). The CD1901 and Lederle strains were used as positive controls. Five canine viruses [rabies virus (ERAGS strain), CAV-1, CAV-2, CPV, and CPIV,] were subjected to the specificity test. Four commercial DAPP vaccines and one negative sample were used for multiplex RT-PCR, which was performed under the conditions described above.
RESULTS
CPEs of the CD1901 and Lederle strain
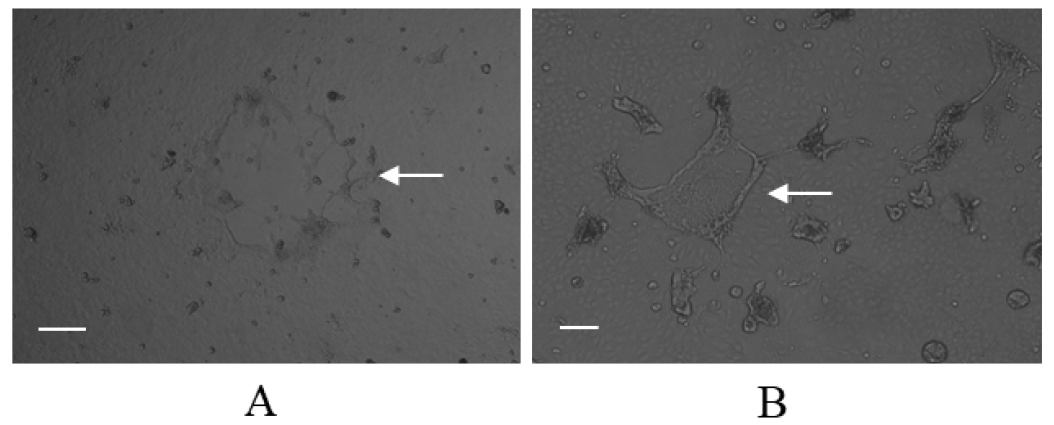
Infection of the Vero/dSLAM cells with the CD1901 and Lederle strains resulted in CPEs showing syncytia formation. As shown in Fig. 3, the CPEs caused by the two CDVs were indistinguishable with the naked eye, nevertheless, the CPE of CD1901 and Lederle strains looked slightly different. The viral titers of the CD1901 and Lederle strains were 106.5 and 105.0 median tissue culture infectious dose (TCID50/ml), respectively. These CDVs were used as positive controls to determine the sensitivity and specificity of multiplex RT-PCR.
Design of primers for multiplex RT-PCR
A common multiplex RT-PCR CDV primer set was designed to detect both the CD1901 and Lederle strains, and the CD1901-F/-R and Lederle-F/-R primer sets were designed to specifically detect the CD1901 and Lederle strains, respectively. Four of the first five nucleotide sequences of the CD1901-R primer were different from those of the Lederle strain, while the first four nucleotide sequences of the Lederle-R primer were different from those of the CD1901 strain. This was due to differences in the starting sites of the reverse primers when amplifying and synthesizing cDNA.
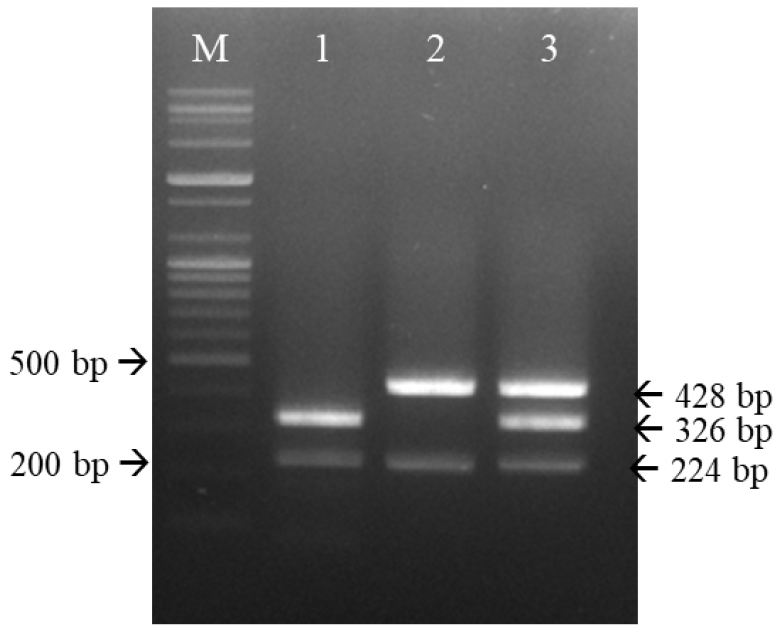
The annealing temperature of multiplex RT-PCR should be set such that the six primers bind optimally to the target DNA. In this study, multiplex RT-PCR was initially carried out at 50°C and 55°C; as shown in Fig. 4, amplification at the latter temperature distinguished the two CDV strains. Thus, all subsequent tests were performed at 55°C.
Sensitivity and specificity of multiplex RT-PCR
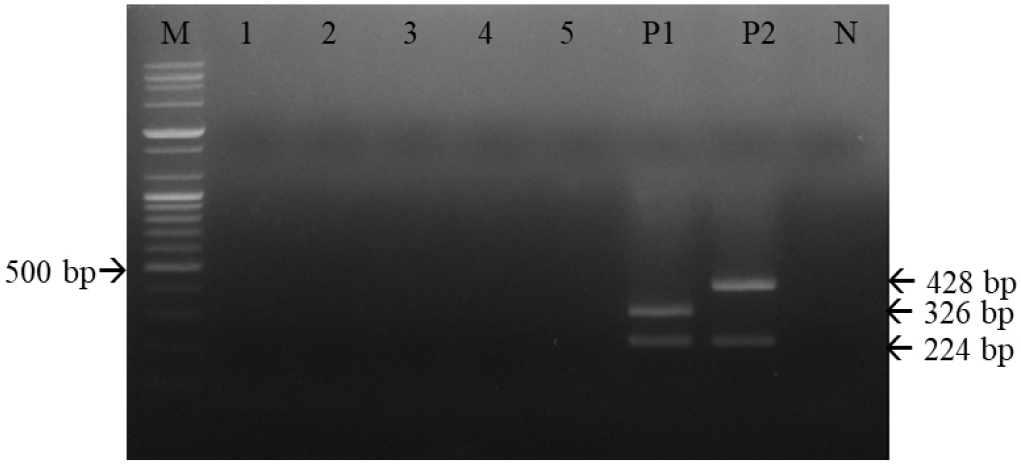
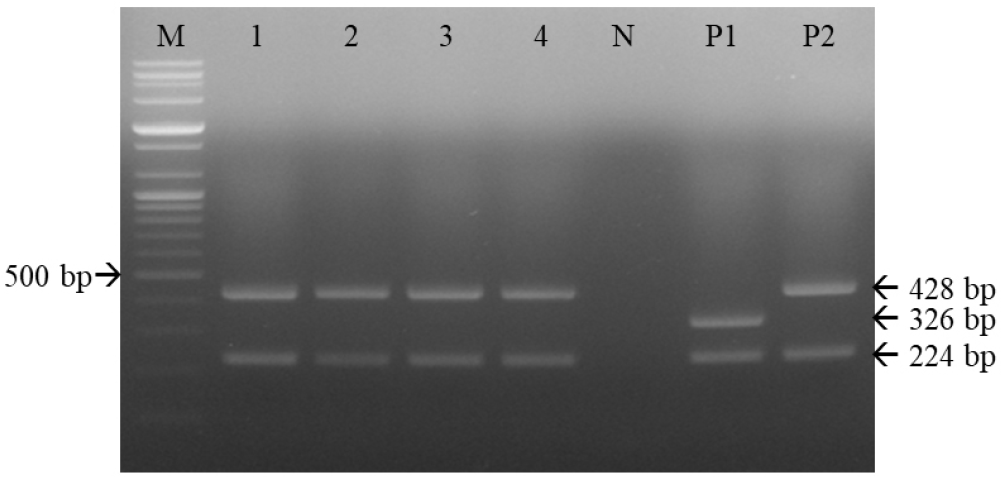
Serial 10-fold dilutions of the CD1901 and Lederle strains were subjected to multiplex RT-PCR amplification. As shown in Table 2 and Fig. 5, the detection limits of multiplex RT-PCR for the CD1901 and Lederle strains were 2.53 and 0.8 TCID50/reaction, respectively. The rabies virus, CAV-1, CAV-2, CPV and CPIV were subjected to multiplex RT-PCR using the three primer sets, and the specificity of the reactions was determined. As shown in Fig. 6, multiplex RT-PCR amplification using the CD1901 and Lederle strains as positive controls yielded only two RT-PCR products (224/346 and 224/428 bp), both of which represented CDV.
Table 2.
Infectious titers of CDV strains detected by multiplex RT-PCR
|
Virus dilution |
Infectious CD1901 titer* |
Multiplex RT-PCR result |
Infectious Lederle titer |
Multiplex RT-PCR result |
|---|---|---|---|---|
| CDV complex/ CD1901 (224/326 bp) |
CDV complex/Lederle (224/428 bp) |
|||
| 10-1 | 2,530 | +/+ | 80 | +/+ |
| 10-2 | 253 | +/+ | 8 | +/+ |
| 10-3 | 25.3 | +/+ | 0.8 | +/+ |
| 10-4 | 2.53 | +/+ | 0.08 | -/- |
| 10-5 | 0.253 | -/- | 0.008 | -/- |
| 10-6 | 0.0253 | -/- | 0.0008 | -/- |
| 10-7 | 0.00253 | -/- | 0.00008 | -/ |
Fig. 5
Sensitivity of the primer sets used for differentiating the CD1901 and Lederle strains. M, 100-bp DNA ladder; 1–7, CD1901 strains 10-1 to 10-7 (A). M, 100-bp DNA ladder; 8–14, Lederle strains 10-1 to 10-7 (B).
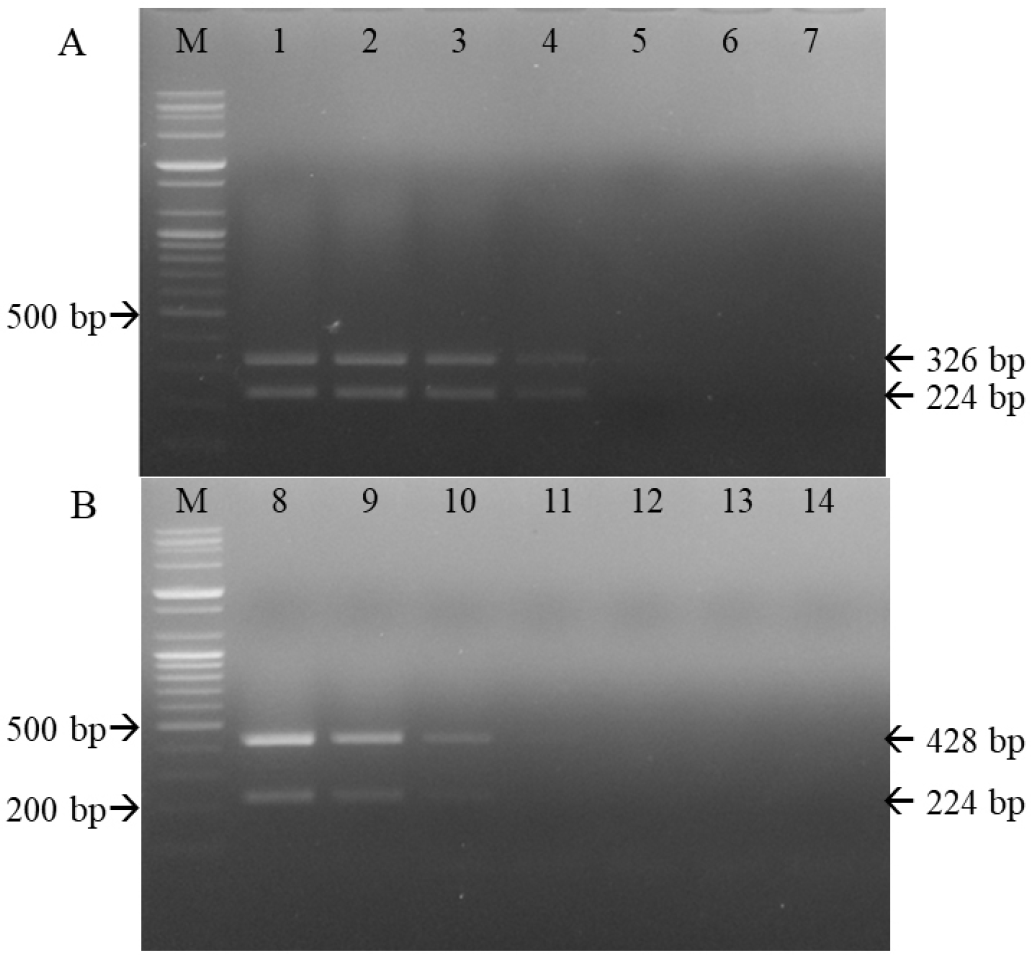
Fig. 6
Specificity of multiplex reverse-transcription polymerase chain reaction (RT-PCR) for canine distemper virus (CDV) strains using the three primer sets. Multiplex RT-PCR effectively detected CDVs; among the five canine viral pathogens, there were only two RT-PCR products, both representing CDV. M, 100-bp DNA ladder; 1, rabies virus; 2, canine parainfluenza virus; 3, canine adenovirus type 1; 4, canine adenovirus type 2; 5, canine parvovirus; P1, CD1901; P2, Lederle strain; N, negative control.
Application of multiplex RT-PCR to commercial DAPP vaccines
One commercial DAPP vaccine containing the Rockborn strain, and three containing the Lederle strain, were subjected to multiplex RT-PCR. As shown in Fig. 7, two DNA bands, of 224 and 428 bp, were identified among these four commercial DAPP vaccines.
DISCUSSION
CDV is a highly contagious pathogen that, along with rabies virus, CPV, and canine influenza viruses, affects dogs and wild raccoon dogs. Modified live virus vaccines containing the Lederle, Rockborn, and Onderstepoort strains and a recombinant canarypox virus expressing the CDV F and H genes have been used to prevent wild CDV infections in canine populations worldwide (13, 14). Recently, the CD1901 strain was isolated from a naturally infected Korean dog using Vero/dSLAM cells (9). Molecular biological analysis revealed many genetic differences in the F gene between the CD1901 and Lederle strains. Therefore, a diagnostic method for differentiating between CD1901 belonging to Asia-1 genotype and the Lederle and Rockborn CDV vaccine strains is needed. In this study, we designed three primer sets and established a multiplex RT-PCR method for simultaneous identification and differentiation of CDV strains via a one-step reaction.
Blood and urine samples, and conjunctival and rectal swabs, can be subjected to CDV diagnosis in dogs. RIDTs only take approximately 10 minutes to complete and are often used in veterinary clinics due to their rapidity (15, 16). However, RIDTs require large amounts of viral antigens. Real-time RT-PCR and reverse-transcription loop-mediated isothermal amplification are the most sensitive diagnostic methods for CDV, with detection limits in the range of 0.1–10 TCID50/ml (12, 17). RT-PCR was able to detect the CDV gene in the blood of two vaccinated Brazilian dogs with asymptomatic canine distemper (18). However, the abovementioned diagnostic methods do not allow for differentiation among CDV genotypes or viral strains. In contrast, multiplex RT-PCR and triplex PCR/RT-PCR have shown efficacy in differentiating CDV or in detecting three pathogens at once (19, 20).
Primer sets targeting conserved genes and generating PCR products of different sizes are important for multiplex RT-PCR (17, 21). In our experiment, the positions of the primer sets used for differentiation and amplification were based on the F and N genes, respectively. The reverse primer used for the CD1901 strain was designed to target nucleotide sequences within the F protein because four of the first five nucleotides differed between the CD1901 and Lederle strains.
When designing reverse primers, the annealing temperature is an important consideration. In a previous study, the annealing temperature for multiplex RT-PCR was set to 51.5°C (19). Using an annealing temperature of 50°C, our multiplex RT-PCR resulted in non-specific amplification (data not shown).
To determine sensitivity, total RNA extracted from the CD1901 and Lederle strains was subjected to multiplex RT-PCR. The sensitivity of the CD1901 primer was 3.16 times lower than that of the Lederle primer set, and the multiplex RT-PCR detection limits for the CD1901 and Lederle strains were 2.53 and 0.8 TCID50/reaction, respectively. The detection limits for CDV using multiplex RT-PCR have been reported in the range of 0.1–100 TCID50/reaction, as stated above (17, 21). In this study, slight differences in sensitivity among the primers may be attributable to the sites targeted, i.e., the N or F gene.
The multiplex RT-PCR performed in this study showed good specificity; no reaction products were found for the rabies virus, CAV-1, CAV-2, CPV and CPIV. CDV was amplified successfully in samples of four commercial DAPP vaccines manufactured in South Korea. However, further studies are required to determine the performance of the multiplex RT-PCR assay for analyzing large numbers of wild CDV-infected samples.
In conclusion, the multiplex RT-PCR used in this study showed high sensitivity and specificity for differentiating between the CD1901 and Lederle CDV strains. Thus, our method can be used for reliable confirmatory diagnosis of CDV infection in suspected animals and for detecting the CD1901 strain in veterinary biological products.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download