Abstract
Background
Methods
Results
Conclusions
Notes
AUTHOR CONTRIBUTIONS
Nam M analyzed the data and wrote the draft; Hur M designed the study and finalized the draft; Yoon S, Park M, and Kim HN participated in data collection; Lee GH and Kim H reviewed the draft. All authors have accepted responsibility for the entire content of this manuscript and approved the final version.
REFERENCES
Fig. 1
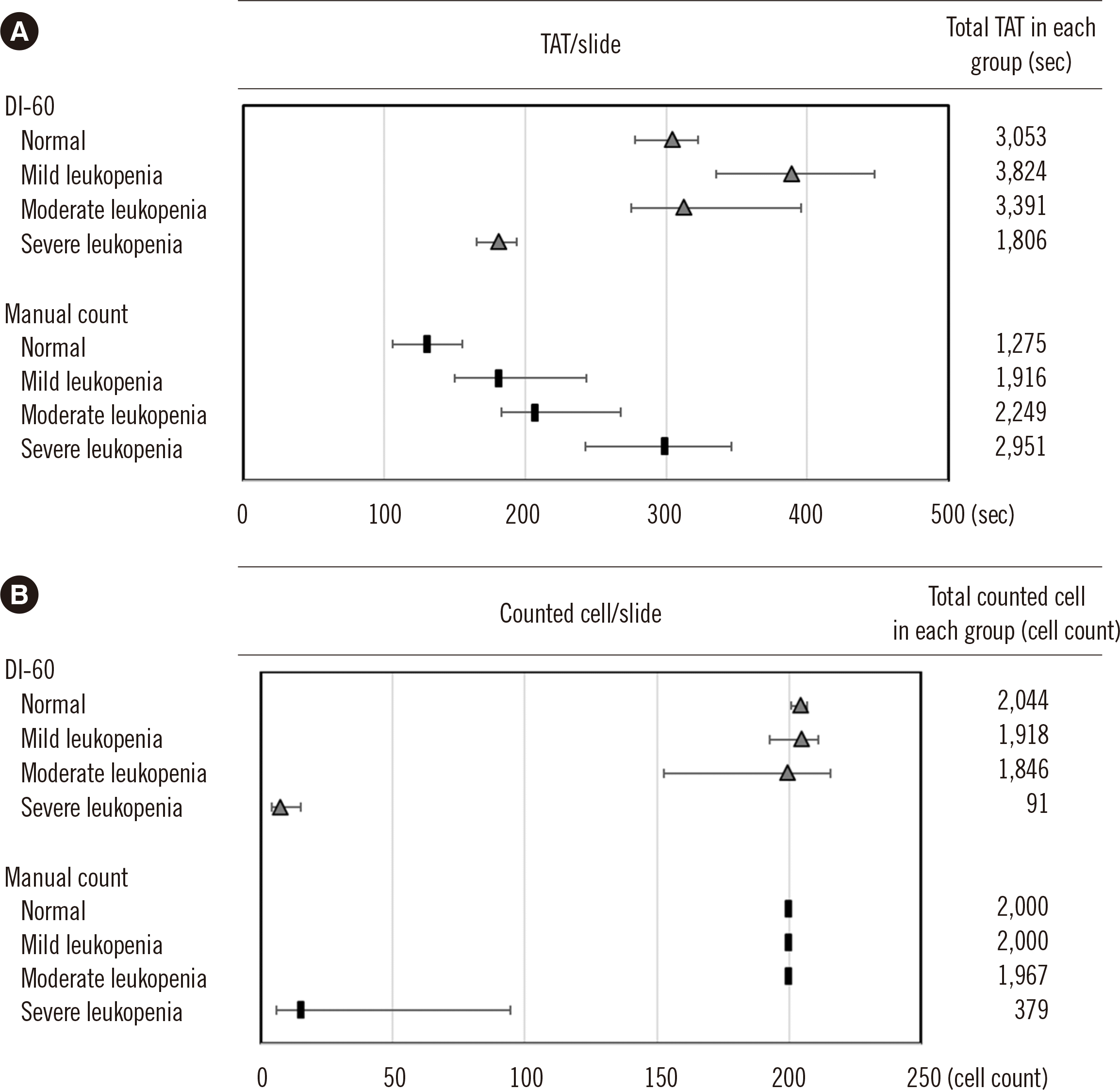
Table 1
| Total (N = 40) | Normal (N = 10) | Mild leukopenia (N = 10) | Moderate leukopenia (N = 10) | Severe leukopenia (N = 10) | |
|---|---|---|---|---|---|
| Age, median (IQR) | 64 (33-76) | 47 (33-71) | 77 (64-78) | 62 (62-73) | 33 (33-65) |
| Male, N (%) | 21 (52.5) | 6 (60) | 6 (60) | 2 (20) | 7 (70) |
| Diagnosis, N (%) | |||||
| AML | 18 (45) | 3 (30) | 1 (10) | 7 (70) | 7 (70) |
| Lymphoma* | 5 (12.5) | - | 3 (30) | 2 (20) | - |
| PCM | 4 (10) | 1 (10) | 1 (10) | - | 2 (20) |
| MDS | 3 (7.5) | - | 3 (30) | - | - |
| ALL | 1 (2.5) | - | - | 1 (10) | - |
| CML | 1 (2.5) | 1 (10) | - | - | - |
| AA | 1 (2.5) | - | - | - | 1 (10) |
| No hematologic diseases† | 7 (17.5) | 5 (50) | 2 (20) | - | - |
| WBC counts, median (IQR) | 2,215 (610-4,863) | 7,010 (5,708-7,798) | 2,840 (2,615-3,400) | 1,710 (1,125-1,930) | 60 (50-190) |
| WBC differentials‡, N (%) | |||||
| Five differentials | 26 (65) | 10 (100) | 2 (20) | 4 (40) | 10 (100) |
| Blasts | 3 (7.5) | - | 3 (30) | - | - |
| Immature granulocytes§ | 8 (20) | - | 5 (50) | 3 (30) | - |
| nRBCs | 1 (2.5) | - | 1 (10) | - | - |
| Lymphocytes, variant form | 9 (22.5) | - | 6 (60) | 3 (30) | - |
*Lymphoma included follicular lymphoma (N=3) and diffuse large B cell lymphoma (N=2); †No hematologic diseases included bone marrow donors (N=2), liver disease (N=2), cardiac disease (N=2), and breast cancer (N=1); ‡The number of WBC differentials included all numbers of five overlapping differentials, blasts, immature granulocytes, nRBC, and variant lymphocytes; §Immature granulocytes included promyelocytes, myelocytes, and metamyelocytes.
Table 2
| Process step | Potential defect | Potential intervention | Consequence | FMEA | ||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| S | O | D | RPN | |||||
| DI-60 analysis | 1. Insert a slide in the instrument | Wrong slide or mechanical error | Repeat | Delay | 4 | 3 | 1 | 12 |
| 2. Scan ideal zone under low power (10 ×) | Mechanical error or poor image | Repeat | Delay | 3 | 2 | 1 | 6 | |
| 3. Acquire images under high power (100 ×) (pre-classification) | Mechanical error | Repeat | Delay | 4 | 1 | 1 | 4 | |
| 4. Verify results | Incorrect verification | Correct | WR | 6 | 2 | 4 | 48 | |
| Total | - | - | - | 70 | ||||
| Manual counting | 1. Place a slide on the microscope stage | Wrong slide or incorrectly labeled slide | Repeat | Delay, WR | 9 | 1 | 6 | 54 |
| 2. Scan ideal zone under low power (100 ×) | Low quality of slide or broken slide | Re-prepare | Delay | 6 | 2 | 1 | 12 | |
| 3. Count cells under high power (1,000 ×) | Incorrect counting | Repeat | Delay, WR | 9 | 2 | 8 | 144 | |
| 4. Record results | Clerical error | Correct | WR | 10 | 2 | 7 | 140 | |
| Total | - | - | - | 350* | ||||
Table 3
| Process step | TAT (min:sec) (median, IQR) | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| Total (N=40) | Normal (N=10) | Mild leukopenia (N=10) | Moderate leukopenia (N=10) | Severe leukopenia (N=10) | ||
| DI-60 analysis | 1. Insert a slide in the instrument | 0:33 (0:32-0:34) | 0:33 (0:33-0:34) | 0:33 (0:32-0:33) | 0:33 (0:32-0:33) | 0:33 (0:33-0:34) |
| 2. Scan ideal zone under low power (10 ×) | 0:18 (0:17-0:19) | 0:18 (0:17-0:19) | 0:18 (0:18-0:20) | 0:18 (0:17-0:18) | 0:19 (0:17-0:19) | |
| 3. Acquire images under high power (100 ×) (pre-classification) | 2:09 (1:58-2:21) | 2:05 (1:58-2:19) | 2:27 (2:08-2:43) | 2:18 (2:08-2:25) | 1:48 (1:37-1:57) | |
| 4. Verification | 1:53 (0:45-2:45) | 2:03 (1:45-2:27) | 3:09 (2:15-4:02) | 2:11 (1:21-3:00) | 0:20 (0:15-0:27) | |
| Total* | 5:00 (2:36-6:06) | 5:05 (4:38-5:23) | 6:29 (5:36-7:28) | 5:13 (4:35-6:36) | 3:02 (2:46-3:14) | |
| Manual counting† | 1. Place a slide on the microscope stage | 0:03 (0:03-0:04) | 0:03 (0:03-0:04) | 0:03 (0:03-0:04) | 0:04 (0:03-0:04) | 0:03 (0:03-0:04) |
| 2. Scan ideal zone under low power (100 ×) | 0:09 (0:05-0:10) | 0:05 (0:04-0:09) | 0:07 (0:05-0:08) | 0:09 (0:06-0:10) | 0:12 (0:10-0:13) | |
| 3. Count cells under high power (1,000 ×) | 2:42 (1:54-3:52) | 1:47 (1:21-2:01) | 2:36 (2:01-3:30) | 3:02 (2:32-3:59) | 4:30 (3:32-5:21) | |
| 4. Record results | 0:18 (0:14-0:20) | 0:17 (0:15-0:20) | 0:19 (0:17-0:23) | 0:18 (0:14-0:21) | 0:11 (0:09-0:21) | |
| Total* | 3:11 (2:18-4:21) | 2:11 (1:46-2:35) | 3:02 (2:30-4:04) | 3:27 (3:03-4:28) | 5:00 (4:01-5:46) | |
The last process step of data transfer to the laboratory information system was excluded for both DI-60 analysis and manual counting.
*Total value indicates median and IQR of total TAT for counting total cells in each slide in total group and each group; †We only compared TAT for one analytical process of one expert in manual counting; if the result of manual counting would have been determined as the mean of the results obtained by the two experts, the TAT of manual counting would have been significantly longer.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download