1. Steele CM, Miller AJ. Sensory input pathways and mechanisms in swallowing: a review. Dysphagia. 2010; 25(4):323–333. PMID:
20814803.
2. Ertekin C. Electrophysiological evaluation of oropharyngeal dysphagia in Parkinson's disease. J Mov Disord. 2014; 7(2):31–56. PMID:
25360228.
3. Park S, Cho JY, Lee BJ, Hwang JM, Lee M, Hwang SY, et al. Effect of the submandibular push exercise using visual feedback from pressure sensor: an electromyography study. Sci Rep. 2020; 10(1):11772. PMID:
32678239.
4. Shaker R, Geenen JE. Management of dysphagia in stroke patients. Gastroenterol Hepatol (N Y). 2011; 7(5):308–332. PMID:
21857832.
5. Finestone HM, Greene-Finestone LS. Rehabilitation medicine: 2. Diagnosis of dysphagia and its nutritional management for stroke patients. CMAJ. 2003; 169(10):1041–1044. PMID:
14609974.
6. Yeom J, Song YS, Lee WK, Oh BM, Han TR, Seo HG. Diagnosis and clinical course of unexplained dysphagia. Ann Rehabil Med. 2016; 40(1):95–101. PMID:
26949675.
7. Chang MC, Park JS, Lee BJ, Park D. Effectiveness of pharmacologic treatment for dysphagia in Parkinson's disease: a narrative review. Neurol Sci. 2021; 42(2):513–519. PMID:
33201362.
8. Yu KJ, Park D. Clinical characteristics of dysphagic stroke patients with salivary aspiration: a STROBE-compliant retrospective study. Medicine (Baltimore). 2019; 98(12):e14977. PMID:
30896670.
9. Lee JT, Park E, Hwang JM, Jung TD, Park D. Machine learning analysis to automatically measure response time of pharyngeal swallowing reflex in videofluoroscopic swallowing study. Sci Rep. 2020; 10(1):14735. PMID:
32895465.
10. Lee JT, Park E, Jung TD. Automatic detection of the pharyngeal phase in raw videos for the videofluoroscopic swallowing study using efficient data collection and 3D convolutional networks †
. Sensors (Basel). 2019; 19(18):3873.
11. Park D, Lee HH, Lee ST, Oh Y, Lee JC, Nam KW, et al. Normal contractile algorithm of swallowing related muscles revealed by needle EMG and its comparison to videofluoroscopic swallowing study and high resolution manometry studies: a preliminary study. J Electromyogr Kinesiol. 2017; 36:81–89. PMID:
28763682.
12. Park D, Oh Y, Ryu JS. Findings of abnormal videofluoroscopic swallowing study identified by high-resolution manometry parameters. Arch Phys Med Rehabil. 2016; 97(3):421–428. PMID:
26505655.
13. Park D, Shin CM, Ryu JS. Effect of different viscosities on pharyngeal pressure during swallowing: a study using high-resolution manometry. Arch Phys Med Rehabil. 2017; 98(3):487–494. PMID:
27523910.
14. Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys. 2019; 29(2):102–127. PMID:
30553609.
15. Ahmed Z, Mohamed K, Zeeshan S, Dong X. Artificial intelligence with multi-functional machine learning platform development for better healthcare and precision medicine. Database (Oxford). 2020; 2020:baaa010. PMID:
32185396.
16. Martin-Harris B, Jones B.. The videofluorographic swallowing study. Phys Med Rehabil Clin N Am. 2008; 19(4):769–785. PMID:
18940640.
17. Pauloski BR, Rademaker AW, Lazarus C, Boeckxstaens G, Kahrilas PJ, Logemann JA. Relationship between manometric and videofluoroscopic measures of swallow function in healthy adults and patients treated for head and neck cancer with various modalities. Dysphagia. 2009; 24(2):196–203. PMID:
18956228.
18. Uhm KE, Yi SH, Chang HJ, Cheon HJ, Kwon JY. Videofluoroscopic swallowing study findings in full-term and preterm infants with Dysphagia. Ann Rehabil Med. 2013; 37(2):175–182. PMID:
23705111.
19. Yamashita R, Nishio M, Do RK, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Imaging. 2018; 9(4):611–629. PMID:
29934920.
20. Tan M, Le QV. EfficientNet: rethinking model scaling for convolutional neural networks. Proc Mach Learn Res. 2019; 97:6105–6114.
21. Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T, et al. Mobilenets: efficient convolutional neural networks for mobile vision applications. arXiv. April 17, 2017. https://arxiv.org/abs/1704.04861.
22. Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. In : Proceedings of the 2016 IEEEE Conference on Computer Vision and Pattern Recognition; 2016 Jun 27-30; Las Vegas, NV, USA. Piscataway, NJ, USA: Institute of Electrical and Electronics Engineers;2016. p. 2818–2826.
23. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In : Proceedings of the 2016 IEEEE Conference on Computer Vision and Pattern Recognition; 2016 Jun 27-30; Las Vegas, NV, USA. Piscataway, NJ, USA: Institute of Electrical and Electronics Engineers;2016. p. 770–778.
24. DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988; 44(3):837–845. PMID:
3203132.
25. Mandrekar JN. Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol. 2010; 5(9):1315–1316. PMID:
20736804.
26. Zhang Z, Coyle JL, Sejdić E. Automatic hyoid bone detection in fluoroscopic images using deep learning. Sci Rep. 2018; 8(1):12310. PMID:
30120314.
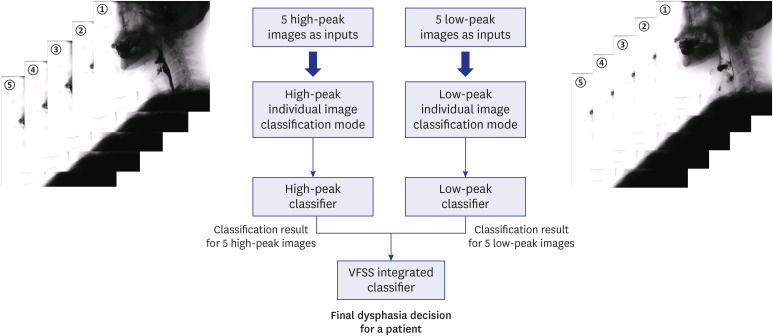
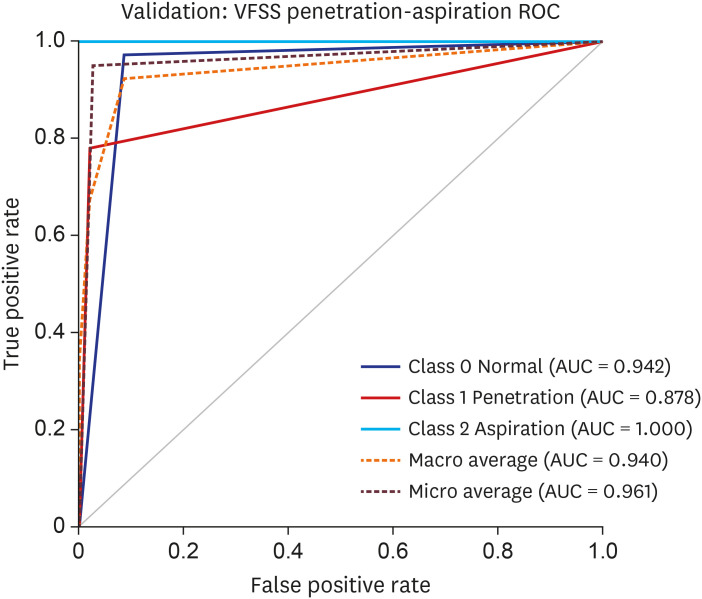
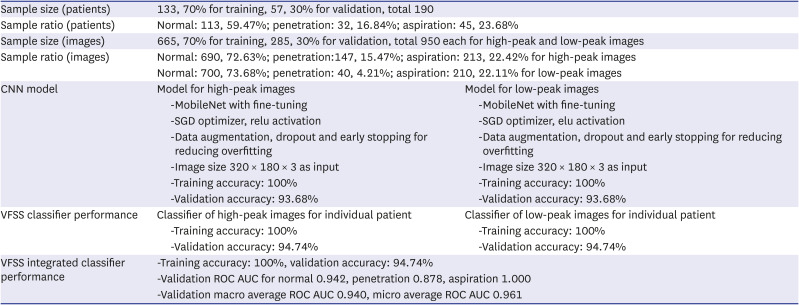
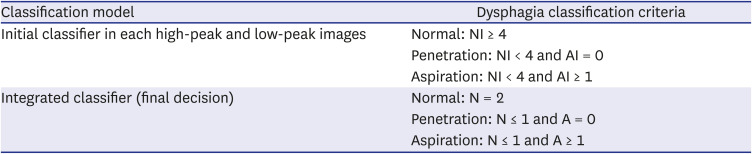




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download