Abstract
Purpose
The purpose of this study was to reveal the clinicopathological characteristics and prognostic implications associated with fibroblast growth factor receptor 1 (FGFR1) amplification in colorectal cancers (CRCs).
Materials and Methods
We measured the copy number of FGFR1 by droplet digital polymerase chain reaction (ddPCR), and analyzed the FGFR1 expression by immunohistochemistry, in 764 surgically resected CRCs (SNUH2007 dataset, 384 CRCs; SNUH Folfox dataset, 380 CRCs).
Results
CRCs with ≥ 3.3 copies of the FGFR1 gene were classified as FGFR1 amplified. FGFR1 amplification was found in 10 of the 384 CRCs (2.6%) in the SNUH2007 dataset, and in 28 of the 380 CRCs (7.4%) in the SNUH Folfox dataset. In the SNUH2007 dataset, there was no association between the FGFR1 copy number status and sex, gross appearance, stage, or differentiation. High FGFR1 expression was associated with female sex and KRAS mutation. At the molecular level, FGFR1 amplification was mutually exclusive with BRAF mutation, microsatellite instability, and MLH1 methylation, in both SNUH2007 and SNUH Folfox datasets. Survival analysis revealed that FGFR1 amplification was associated with significantly worse clinical outcome compared with no FGFR1 amplification, in both SNUH2007 and SNUH Folfox datasets. Within the SNUH2007 dataset, CRC patients with high FGFR1 expression had an inferior progression-free survival compared with those with low FGFR1 expression. The FGFR inhibitor, PD173074, repressed the proliferation of a CRC cell line overexpressing FGFR1, but not of cells with FGFR1 amplification.
Colorectal cancers (CRCs) are the third-most commonly diagnosed malignancy in the United States and South Korea [1,2]. CRCs develop as a result of the accumulation of genetic and epigenetic alterations, and recent comprehensive genomic and epigenomic analyses have greatly increased our understanding of the genomic alterations involved in CRCs [3]. Although alternative carcinogenic pathways, such as microsatellite instability (MSI) and CpG island methylator phenotype (CIMP), have been described, the vast majority of CRCs (approximately 70%) exhibit chromosomal instability (CIN), which is characterized by the accumulation of somatic mutations and copy number alterations (CNAs) [4]. Functionally, CRCs are dominated by somatic mutations, rather than CNAs [5]. However, several genomic regions show significant focal amplification or deletion of CRCs [3].
Fibroblast growth factor receptor 1 (FGFR1) gene encodes a transmembrane receptor tyrosine kinase and maps to chromosome 8p11 [6]. The FGFR family includes four receptor tyrosine kinases, FGFR1-4, and its structural variability is derived from alternative splicing [6]. FGFR ligands, fibroblast growth factors (FGFs) activate downstream signaling pathways including mitogen-activated protein kinase and phosphoinositide-3-kinase/AKT [6]. FGF signaling promotes cell proliferation, cell survival, and angiogenesis, resulting in tumor development [6]. Amplification of the FGFR1 gene is reported in estrogen receptor (ER)–positive breast cancers, lung cancers, esophageal cancers, and bladder cancers. Recently, FGFR1 amplification has been suggested to be associated with poor prognosis in various types of cancers, including squamous cell carcinoma of the lung and esophagus, and ERpositive breast cancers [7-9].
Fluorescence in situ hybridization (FISH) is the gold-standard method for the evaluation of CNAs in clinical oncology [10]. However, FISH has several disadvantages, such as the requirement of a fluorescence microscope and dark room, subjective measurement of fluorescence signals, spontaneous weakening of fluorescence with time, and high cost. Chromogenic in situ hybridization (CISH) and silver-enhanced in situ hybridization (SISH) have been developed to overcome the fluorescence-associated limitations of FISH [11,12]. However, the subjectivity associated with visual scoring remains an issue for CISH and SISH. Although quantitative measurement of nucleic acids using polymerase chain reaction (PCR) has been considered as an alternative method for CNA analysis, quantitative real-time PCR is not widely used in clinical practice, owing to problems related to reproducibility. Recently, water-oil emulsion droplet technology-based third-generation PCR technology, termed droplet digital PCR (ddPCR), has been developed. The ddPCR technique offers a number of advantages for both detection and quantification of nucleic acids, including the capacity to measure fold-change and detect rare variants [13]. Compared with real-time quantitative PCR, ddPCR is more robust technique less prone to PCR inhibition. Moreover, ddPCR offers improved day-to-day reproducibility without requiring a standard curve of reference material. Take advantage of high accuracy to detect rare variants, interest in ddPCR is rapidly rising in the field of liquid biopsy which detect circulating tumor DNA [13]. Therefore, ddPCR could be a complementary method to evaluate CNA in clinical oncology.
FGFR1 amplification is found in 2% to 5% of CRCs [14]. However, the clinicopathologic characteristics and prognostic implications associated with FGFR1 amplification in CRCs are not well established because of the scarcity of this alteration. In this study, we evaluated the FGFR1 copy number in CRCs using ddPCR and evaluated its association with clinicopathologic characteristics and prognostic implications.
A total of 538 patients with CRCs underwent surgical treatment at Seoul National University Hospital between January 2007 to December 2007, consecutively. After excluding the patients who refused to participate in the molecular study, as well as those who had non-invasive cancers, a history of neo-adjuvant treatment, familial adenomatous polyposis, multiple tumors, or recurrent tumors [15], 384 patients were eligible and willing to participate, all of whom were included in the clinicopathological and molecular analysis.
We obtained tissues from 405 patients with high-risk stage II or III CRC who received adjuvant FOLFOX treatment from August 2005 to December 2011. After the elimination of patients who fulfilled the exclusion criteria for SNUH2007 dataset, 380 patients were selected for the validation set.
DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) tumor tissues. The representative tumor areas were delineated under a light microscope on 10 serial unstained slides of tumor blocks. DNA extraction was performed after macro-dissection using ZR FFPE DNA MiniPrep kit (Zymo Research, Orange, CA) according to the manufacturer’s protocol.
ddPCR (QX200, Bio-Rad, Hercules, CA) was used in this study. Each sample was partitioned into 20,000 droplets, with target and background (reference) DNA randomly, but not uniformly, distributed among the droplets [16]. The following primers were used for ddPCR: FGFR1 Hs02882334_cn (Life Technologies), RPPH1-ddF 5′-GCGGATGCCTCCTTTGC-3′, RPPH1-ddR 5′-ACCTCACCTCAGCCATTGAACT-3′, RPPH1-HEX HEX-CTTGGAACAGACTCACGGCCAGCG-BHQ1. The reactions were performed in 20 μL reaction mixtures that comprised up to 100 ng of the extracted DNA (5 μL), 2× ddPCR supermix for probe (10 μL), the FGFR1 primer (1 μL), the RPPH1 primer (1.8 μL), RPPH1-HEX (0.5 μL), HindIII (0.3 μL), and deionized distilled water (1.4 μL). The emulsified PCR reactions were run in 96-well plates on a thermal cycler. The plates were incubated at 95& for 10 minutes, followed by 50 cycles of 94& for 30 seconds, 60& for 50 seconds, and 72& for 30 seconds, and finally, a 10-minute incubation at 98&. The number of droplets positive for FGFR1 and/or RPPH1 in each plate was assessed on a Bio-Rad QX200 droplet reader using the QuantaSoft v1.7 software (Bio-Rad). FGFR1 gene copy number determined by ddPCR was defined as 2× FGFR1/RPPH1. The cut-off for classifying samples as FGFR1 amplified was set as ≥ 3.3 copies/cell (FGFR1/RPPH ≥ 1.65), using maximally selected chi-square statistics.
Using histological examination, we marked the tumor areas in the tissue section samples from each patient. Pairs of 2-mm core tumor tissue samples were subsequently extracted from each paraffin-embedded formalin tissue sample (donor block) and arranged in a new tissue microarray block using a trephine apparatus [15]. To analyze the FGFR1 protein expression, we performed immunohistochemical staining with rabbit monoclonal anti-FGFR1 antibody (1:50, clone EPR806Y, Abcam, Cambridge, MA). We classified the staining intensity of cytoplasmic or membranous FGFR1 staining as 0 (no stain), 1 (mild stain), 2 (moderate stain), or 3 (strong stain). The level of FGFR1 expression was subsequently categorized as low (FGFR1 intensity 0 and 1) or high (FGFR1 intensity 2 and 3).
Direct sequencing of KRAS codons 12 and 13 and allelespecific PCR for BRAF codon 600 were performed as previously described [15]. The microsatellite status of each tumor was determined by the evaluation of five microsatellite markers: D2S123, D5S346, D17S250, BAT25, and BAT26. The MSI status was classified as follows: MSI-high (instability at ≥ 2 microsatellite markers), MSI-low (instability at 1 marker), or microsatellite stable (no instability) [17].
After sodium bisulfite conversion of DNA using an EZ DNA methylation kit (Zymo Research), the methylation status was quantified at the following eight CIMP markers using MethyLight assay, as previously described: CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1 [15]. The CIMP status was defined based on the number of methylated markers as follows: CIMP-negative (CIMPN, 0-4 methylated markers), CIMP-positive 1 (CIMP-P1, 5-6 markers), or CIMP-positive 2 (CIMP-P2, 7-8 markers) [15].
Five CRC cell lines, with FGFR1 amplification (SNU-C1), high FGFR1 expression (SNU-283, SW620 and HCT116), and no FGFR1 amplification and low FGFR1 expression (SNU-81) were grown for 72 hours in RPMI-1640 or Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, at 37& under a humidified 5% CO2 atmosphere. The FGFR1 copy number status in these five cell lines was confirmed by ddPCR, while the FGFR1 expression was confirmed by Western blot. The effect of FGFR inhibitor, PD173074, on cell proliferation was determined using the WST-1 assay in SNU-C1, and 3-(4,5-dimethylthiazol-2-yl)-2-5-diphenyltetrazolium bromide (MTT) cell viability assay in the other cell lines [18]. For the WST-1 assay, after the indicated treatment, 20 μL of WST-1 solution was added per well and the cells were incubated for 4 hours at 37&. The optical density (OD) was measured at a wavelength of 440 nm. For the MTT assay, after the indicated treatment, 50 μL of MTT reagent (5 mg/mL) was added per well and the cells were incubated for 4 hours at 37&. Next, 150 μL of dimethyl sulfoxide was added to lyse the cells and solubilize the colored crystals. Plates were then incubated for 10 minutes at 37&, and OD was measured using a microplate reader at a wavelength of 570 nm. Halfmaximal inhibitory concentrations (IC50) were calculated using GraphPad Prism 8 (San Diego, CA).
Molecular characteristics of The Cancer Genome Atlas (TCGA) COADREAD dataset, including CNA and mRNA expression status of FGFR1 and BRAFmutations, were obtained from cBioPortal for cancer genomics (http://cbioportal.org). MSI, MLH1 methylation, CIMP status, and survival data for the TCGA COADREAD dataset were obtained from previous studies [19,20].
For the comparison between CRCs with and without FGFR1 amplification, we compared the genome-wide DNA methylation and transcriptome data for colon and rectum adenocarcinoma (COAD, READ), breast invasive carcinoma (BRCA), and lung squamous cell carcinoma (LUSC) from the TCGA dataset. To identify differentially methylated CpGs (dmCp-Gs), the methylation status of each probe was examined using Empirical Bayes moderated t test using the limma package in R/Bioconductor. The threshold for the identification of dmCpG was Benjamini and Hochberg (BH)-adjusted p < 0.05. To identify differentially expressed genes (DEGs), the normalized RNA sequencing by expectation maximization value of each gene was examined using the DESeq2 package in R/Bioconductor. The threshold for the identification of DEG was |log2(fold change)| ≥ 1 and BH-adjusted p < 0.05.
Categorical variables were compared using the chi-square test, Fisher exact test, or ANOVA test, as appropriate. For ordinal variables, Wilcoxon’s rank-sum test was performed. Progression-free survival (PFS) and disease-free survival (DFS) were calculated using the log-rank test with Kaplan-Meier curve. Hazard ratios were calculated using the Cox proportional hazard model. The assumption of proportional hazards was verified by plotting the log(–log(S(t)) against the time of the study. For the modeling, all the variables associated with PFS or DFS with a p < 0.10 were entered into an initial model, after which, the number of variables was reduced by backward elimination. All statistical tests were twosided, and statistical significance was defined as p < 0.05.
This study was approved by the Institutional Review Board (IRB) of the Seoul National University Hospital (No. H-1601-027-733). Due to retrospective nature of this study, informed consent was waived by IRB. This study was performed in accordance with the recommendations of the Declaration of Helsinki for biomedical research involving human subjects.
To compare the results of ddPCR with those of FISH for copy number analysis, we measured the FGFR1 gene copy number using ddPCR in 20 FFPE CRC samples, for which the presence of FGFR1 gene copy had already been established by FISH [21]. The results of ddPCR and FISH results were strongly correlated (Spearman’s rho, 0.686; p=0.001) (S1 Fig.). Based on a cut-off of ≥ 3.3 FGFR1 copies/cell for ddPCR and ≥ 2 FGFR1/CEP8 for FISH, ddPCR and FISH showed a strong concordance for FGFR1 amplification (κ=0.875) (S2 Table).
A total of 384 patients with CRC (median age, 63 years; min-max, 28 to 84 years) were included in the study from the SNUH2007 dataset. The male-to-female ratio was 1.42:1 (225 males and 159 females). The tumor location was the proximal colon (proximal to the splenic flexure) in 77 patients, the distal colon in 143 patients, and the rectum in 164 patients. The median follow-up duration was 68.8 months, and 291 patients received 5-fluorouracil based adjuvant chemotherapy.
From the SNUH Folfox dataset, a total of 380 patients with high-risk stage II or stage III CRC, who received adjuvant FOLFOX (median age, 60; min-max, 29 to 78) were selected. The male-to- female ratio was 1.57 (232 males and 148 females). The tumor location was the proximal colon in 131 patients, the distal colon in 210 patients and the rectum in 39 patients. The median follow-up duration was 71.7 months.
In the SNUH2007 dataset, the median FGFR1 gene copy number was 1.10 (min-max, 0.15 to 15.00). Based on a cut-off value of 3.3, 10 patients (2.6%) showed FGFR1 amplification. Immunohistochemical FGFR1 expression data were obtained for 382 of the 384 patients. Among these 382 patients, 50 (13.1%) showed moderate to strong FGFR1 expression, whereas 332 patients (86.9%) showed no or weak FGFR1 expression (S3 Fig.).
In the SNUH Folfox dataset, the median FGFR1 gene copy number was 1.71 (min-max, 0.35 to 19.20), and 28 patients (7.4%) showed FGFR1 amplification. Immunohistochemical FGFR1 expression data were obtained for 351 of the 380 patients. Among these 351 patients, 58 (16.5%) showed moderate to strong FGFR1 expression, whereas 293 patients (83.5%) showed no or weak FGFR1 expression. There was no significant correlation between FGFR1 copy number and FGFR1 expression in either the SNUH2007 dataset (p for ANOVA=0.789) or the SNUH Folfox dataset (p for ANOVA=0.889) (S4A and S4B Fig.). Moreover, there was no significant correlation between the copy number and mRNA expression of FGFR1 in the TCGA COADREAD dataset (p for ANOVA=0.187) (S4C Fig.).
As the positive correlation between FGFR1 amplification and FGFR1 expression is well addressed in breast cancer and squamous cell carcinoma of the lung [7,22], we compared the genome-wide DNA methylation data and transcriptome data of TCGA COADREAD, BRCA, and LUSC datasets (S5 Fig.). In transcriptome analysis, TCGA COADREAD dataset showed strong positive correlation of FGFR1mRNA expression with that of three genes (AKAP12, GEM, and NOTCH4). However, in TCGA BRCA and LUSC dataset, these genes did not show strong correlation with FGFR1 expression. In DNA methylation analysis, TCGA COADREAD dataset showed a hypomethylation of CpGs located in the gene body which correspond to AKAP12, GEM, and NOTCH4 (cg09846917, cg22885-024, and cg26793289, respectively) in cases with FGFR1 amplification compared with cases without FGFR1 amplification. However, the corresponding CpGs were not hypo-methylated in FGFR1 amplified cases compared with non- amplified FGFR1 in the TCGA BRCA dataset and TCGA LUSC dataset.
To demonstrate the clinicopathological characteristics of CRCs with FGFR1 amplification or high FGFR1 expression in an unbiased manner, we performed exploratory analysis for the SNUH2007 dataset, which consisted of consecutively resected primary CRCs (Table 1). In the SNUH2007 dataset, CRCs with FGFR1 amplification were marginally associated with a lower age of onset (p=0.048). CRCs with high FGFR1 expression were more frequent in female patients (p=0.004), and patients with a KRAS mutation (p=0.030), compared with CRCs with low FGFR1 expression. At the molecular level, FGFR1 amplification was mutually exclusive with BRAF mutation, MSI-high, MLH1 methylation, and CIMPP2 (Fig. 1A). These mutually exclusive patterns were consistently observed in the SNUH Folfox dataset and TCGA COADREAD dataset (Fig. 1B and C). The clinicopathological characteristics of CRCs with FGFR1 amplification or high FGFR1 expression in the SNUH Folfox dataset are summarized in S6 Table.
Univariate survival analysis revealed that in SNUH2007 dataset, FGFR1 amplification and high FGFR1 expression were associated with inferior 5-year PFS (p=0.008 for FGFR1 amplification and p=0.031 for high FGFR1 expression) compared with CRCs with no FGFR1 amplification and low FGFR1 expression, respectively (Fig. 2A, S7A Fig.). Because of the scarcity of FGFR1 amplification in the SNUH2007 dataset, we could not perform a multivariate survival analysis using these data.
To validate the prognostic value of FGFR1 amplification and high FGFR1 expression in an independent dataset, we performed survival analysis in the SNUH Folfox dataset, which was controlled for stage and adjuvant chemotherapy. The univariate survival analysis revealed that patients with FGFR1 amplification exhibited marginally worse 5-year DFS compared with those without amplification (p=0.030) (Fig. 2B). However, there was no significant difference in the 5-year DFS between CRC patients with high vs. low FGFR1 expression (p=0.404) (S7B Fig.). Multivariate survival analysis established, FGFR1 amplification as a prognostic marker for poor 5-year DFS, independent of KRAS mutations, lymphovascular invasion, and gross appearance (hazard ratio, 2.22; 95% confidence interval, 1.04 to 4.77; p=0.040) (Table 2). In the TCGA COADREAD dataset, patients with FGFR1 amplification showed a tendency of poor overall survival, although this was not statistically significant (p=0.300) (Fig. 2C). Moreover, in the TCGA COADREAD dataset, the clinical outcome did not differ with FGFR1 mRNA expression for any of the cut-offs (data not shown).
To investigate whether FGFR1 amplification or high FGFR1 expression is a predictive marker for anti-FGFR drug sensitivity, we performed a cell proliferation assay using the FGFR1 inhibitor PD173074, in CRC cell lines with FGFR1 amplification (SNU-C1), high FGFR1 expression (SNU-283, SW620, and HCT116), and one cell line with no FGFR1 amplification and low FGFR1 expression (SNU-81) [14]. In the cell proliferation assays, cell lines with FGFR1 high-expression (SNU-283, SW620, and HCT116) showed a lower IC50 for PD173074 compared with FGFR1 amplified cell line (SNUC1) (Fig. 3).
ddPCR is a third-generation PCR technology that enables absolute measurement of nucleic acid concentrations, based on limiting dilution, end-point PCR, and Poisson statistics [13]. In ddPCR, the target DNA molecules are partitioned into 15,000-20,000 lipid droplets. After amplification to the terminal plateau phase of PCR, the droplets containing one or more templates yield positive end-points, whereas those without a template remain negative. The number of target DNA molecules present can be calculated from the fraction of positive end-point reactions, using Poisson statistics with the following equation: λ=-ln(1–P), where λ is the average number of target DNA molecules per replicate reaction and P is the fraction of positive end-point reactions [23]. Recently, a series of studies successfully detected HER2 amplification in FFPE samples and plasma DNA from patients with breast and gastric cancer using ddPCR [16,24].
CNA analysis using macro-dissected samples leads to an underestimation of copy number in FGFR1-amplified cancers. As the median tumor cell purity of CRC tissues obtained by manual macro-dissection is approximately 65%, a cut-off of 3.3 for FGFR1 amplification in ddPCR is equivalent to an FGFR1/RPPH1 ratio ≥ 2.0, as measured by FISH (0.65×2.0 FGFR1/RPPH1 in tumor cells+0.35×1.0 FGFR1/RPPH1 in normal cells=3.3/2). An FGFR1/CEP8 ratio ≥ 2 is the accepted value of a cut-off for FGFR1 amplification in various type of cancers [8,25]. Therefore, a cut-off of 3.3 for FGFR1 amplification in macro-dissected samples could be acceptable for the evaluation of FGFR1 amplification.
Little is known about the clinicopathological characteristics of CRCs with FGFR1 gene amplification or FGFR1 highexpression. In the present study, CRCs with FGFR1 amplification were marginally associated with a younger age at diagnosis in the SNUH2007 dataset. The TNM stage and tumor differentiation were not significantly different between the FGFR1-amplified group and the non-amplified group. Molecular analysis revealed that FGFR1 amplification was mutually exclusive with BRAF mutation, MSI-high, MLH1 methylation, and CIMP-P2, in both SNUH2007 and SNUH Folfox datasets. Congruently, this mutual exclusivity was observed in the TCGA COADREAD dataset as well. These results suggested that in CRCs, FGFR1 amplification occurs in the context of CIN, rather than MSI or CIMP.
Unlike breast cancers or squamous cell carcinoma of the lung, which show a positive correlation between FGFR1 amplification and FGFR1 mRNA expression, FGFR1 amplification was not associated with FGFR1 mRNA or FGFR1 protein overexpression in CRCs [7,14,22]. Analysis of genome-wide DNA methylation and transcriptome data revealed that in FGFR1-amplified cases in the TCGA COADREAD dataset, three of the genes co-expressed with FGFR1 (AKAP-12, GEM, and NOTCH4) showed a hypomethylation of CpG sites located in their gene body, compared with the cases with non-amplified FGFR1. In the TCGA BRCA and LUSC datasets, the methylation of these CpG sites was not different between the cases with amplified vs. non-amplified FGFR1. Even though the exact relationship of these genes with FGFR1 is not well known, reduced expression of these genes could be a clue to the absence of positive correlation between FGFR1 amplification and FGFR1 mRNA or FGFR1 protein expression in CRCs, because gene body methylation is associated with reduced expression of corresponding genes [26].
Sato et al. [27] evaluated the FGFR1 mRNA expression using quantitative real-time PCR in 202 CRC patients and reported that CRCs with FGFR1 mRNA high-expression were associated with liver metastasis. Goke et al. [14] evaluated the pFGFR1 expression by immunohistochemistry in 99 CRC patients and reported that CRCs showing membranous pFGFR1 expression of cytoplasmic FGFR1 expression were frequently observed in male patients. Moreover, they found that the nuclear localization of pFGFR1 was associated with lymphatic invasion and angioinvasion [14]. When we evaluated the FGFR1 expression by immunohistochemistry in 382 primary CRCs from the SNUH2007 dataset, high FGFR1 expression was frequently observed in female patients. Moreover, there was no significant difference in the TNM stage between CRCs with high vs. low FGFR1 expression in the SNUH2007 dataset.
The prognostic value of FGFR1 amplification in CRCs was not well documented due to its low prevalence. In the present study, CRCs with FGFR1 amplification was associated with a poor 5-year PFS in consecutively resected CRC datasets. In the TCGA COADREAD dataset, FGFR1 amplification showed a tendency of poor overall survival. Moreover, FGFR1 amplification was an independent marker of poor 5-year DFS in patients treated with the Folfox regimen, which is a standard adjuvant combination chemotherapy for high-risk stage II or stage III CRCs. The prognostic value of FGFR1 amplification should be validated in large prospective cohorts.
Several pharmacological agents have been developed to inhibit FGFR activity. These include multi-target receptor tyrosine kinase inhibitors with efficacy against vascular endothelial growth factor receptor, platelet-derived growth factor receptor, and FGFR, as well as selective FGFR inhibitors, which suppress FGFR1-3 [28]. Non-selective broadspectrum receptor tyrosine kinase inhibitors exhibit only a modest bioactivity against FGFR and elicit a wide-spectrum of off-target effects. Selective FGFR inhibitors such as AZD-4547 and BGJ398 have tolerable side effects; however, they did not fulfill the pre-specified efficacy end-point in phase 1 clinical trials [29]. Furthermore, it remains controversial whether FGFR1 amplification or overexpression is an appropriate biomarker for anti-FGFR therapy. Weiss et al. [30] showed that a selective FGFR inhibitor, PD173074, inhibited the growth and induced apoptosis in lung cancer cells with FGFR1 amp-lification. However, in our present study, the lowest IC50 for PD173074 was observed in the HCT116 cell line, which exhi-bits high FGFR1 expression. Furthermore, BGJ398 and AZD4547 are known to have a strong anti-proliferative effect in CRC cell lines with high FGFR1 expression [14,18]. Therefore, proper selection of a biomarker that can predict the res-ponses to FGFR-targeted therapy is muchneeded.
Our present study has several limitations. First, it is a retrospective study, carried out within a single institution. Second, even though poor clinical outcomes for FGFR1 amplified CRCs were consistently found in each dataset, the validation of the prognostic value of FGFR1 amplification is still limited, due to the different clinical settings of each dataset. To validate the results of the present study, external validation using an independent cohort is required.
In conclusion, FGFR1 amplification could be a prognostic indicator of poor clinical outcome in CRCs. Evaluation of somatic CNA using ddPCR is a viable alternative diagnostic method to FISH for clinical diagnostics.
Electronic Supplementary Material
Supplementary materials are available at Cancer Research and Treatment website (https://www.e-crt.org).
ACKNOWLEDGMENTS
This study was supported by grant from the SNUH Research Fund (04-2017-0830), grants from the National Research Foundation (NRF) funded by the Korean Ministry of Science, ICT and Future Planning (2011-0030049 and 2016M3A9B6026921), a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute funded by the Korean Ministry of Health and Welfare (HI14C1277).
References
2. Jung KW, Won YJ, Kong HJ, Lee ES. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2016. Cancer Res Treat. 2019; 51:417–30.

3. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012; 487:330–7.
5. Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013; 45:1127–33.

6. Dienstmann R, Rodon J, Prat A, Perez-Garcia J, Adamo B, Felip E, et al. Genomic aberrations in the FGFR pathway: opportunities for targeted therapies in solid tumors. Ann Oncol. 2014; 25:552–63.

7. Kim HR, Kim DJ, Kang DR, Lee JG, Lim SM, Lee CY, et al. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in patients with resected squamous cell lung cancer. J Clin Oncol. 2013; 31:731–7.

8. Kim HS, Lee SE, Bae YS, Kim DJ, Lee CG, Hur J, et al. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival in patients with resected esophageal squamous cell carcinoma. Oncotarget. 2015; 6:2562–72.

9. Elbauomy Elsheikh S, Green AR, Lambros MB, Turner NC, Grainge MJ, Powe D, et al. FGFR1 amplification in breast carcinomas: a chromogenic in situ hybridisation analysis. Breast Cancer Res. 2007; 9:R23.

10. Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013; 31:3997–4013.

11. Fritzsche FR, Bode PK, Moch H, Kristiansen G, Varga Z, Bode B. Determination of the Her-2/neu gene amplification status in cytologic breast cancer specimens using automated silverenhanced in-situ hybridization (SISH). Am J Surg Pathol. 2010; 34:1180–5.

12. Yoo SB, Lee HJ, Park JO, Choe G, Chung DH, Seo JW, et al. Reliability of chromogenic in situ hybridization for epidermal growth factor receptor gene copy number detection in nonsmall-cell lung carcinomas: a comparison with fluorescence in situ hybridization study. Lung Cancer. 2010; 67:301–5.

13. Huggett JF, Cowen S, Foy CA. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem. 2015; 61:79–88.

14. Goke F, Goke A, von Massenhausen A, Franzen A, Sharma R, Kirsten R, et al. Fibroblast growth factor receptor 1 as a putative therapy target in colorectal cancer. Digestion. 2013; 88:172–81.

15. Bae JM, Kim JH, Kwak Y, Lee DW, Cha Y, Wen X, et al. Distinct clinical outcomes of two CIMP-positive colorectal cancer subtypes based on a revised CIMP classification system. Br J Cancer. 2017; 116:1012–20.

16. Kinugasa H, Nouso K, Tanaka T, Miyahara K, Morimoto Y, Dohi C, et al. Droplet digital PCR measurement of HER2 in patients with gastric cancer. Br J Cancer. 2015; 112:1652–5.
17. Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003; 349:247–57.

18. Yao TJ, Zhu JH, Peng DF, Cui Z, Zhang C, Lu PH. AZD-4547 exerts potent cytostatic and cytotoxic activities against fibroblast growth factor receptor (FGFR)-expressing colorectal cancer cells. Tumour Biol. 2015; 36:5641–8.

19. Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018; 173:400–16.
20. Liu Y, Sethi NS, Hinoue T, Schneider BG, Cherniack AD, Sanchez-Vega F, et al. Comparative molecular analysis of gastrointestinal adenocarcinomas. Cancer Cell. 2018; 33:721–35.
21. Kwak Y, Nam SK, Seo AN, Kim DW, Kang SB, Kim WH, et al. Fibroblast growth factor receptor 1 gene copy number and mRNA expression in primary colorectal cancer and its clinicopathologic correlation. Pathobiology. 2015; 82:76–83.

22. Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010; 70:2085–94.
23. Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011; 83:8604–10.

24. Zhu Y, Lu D, Lira ME, Xu Q, Du Y, Xiong J, et al. Droplet digital polymerase chain reaction detection of HER2 amplification in formalin fixed paraffin embedded breast and gastric carcinoma samples. Exp Mol Pathol. 2016; 100:287–93.
25. Koole K, Brunen D, van Kempen PM, Noorlag R, de Bree R, Lieftink C, et al. FGFR1 is a potential prognostic biomarker and therapeutic target in head and neck squamous cell carcinoma. Clin Cancer Res. 2016; 22:3884–93.
26. Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014; 26:577–90.

27. Sato T, Oshima T, Yoshihara K, Yamamoto N, Yamada R, Nagano Y, et al. Overexpression of the fibroblast growth factor receptor-1 gene correlates with liver metastasis in colorectal cancer. Oncol Rep. 2009; 21:211–6.

28. Shaw AT, Hsu PP, Awad MM, Engelman JA. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer. 2013; 13:772–87.

Fig. 1.
Mutual exclusivity of fibroblast growth factor receptor 1 (FGFR1) amplification with BRAF mutation, microsatellite instability, MLH1 methylation, and CpG island methylator phenotype (CIMP). (A) SNUH2007 dataset. (B) SNUH Folfox dataset. (C) The Cancer Genome Atlas (TCGA) COADREAD dataset. MSS, microsatellite stable; MSI-L, microsatellite instability low; MSI-H, microsatellite instability high.
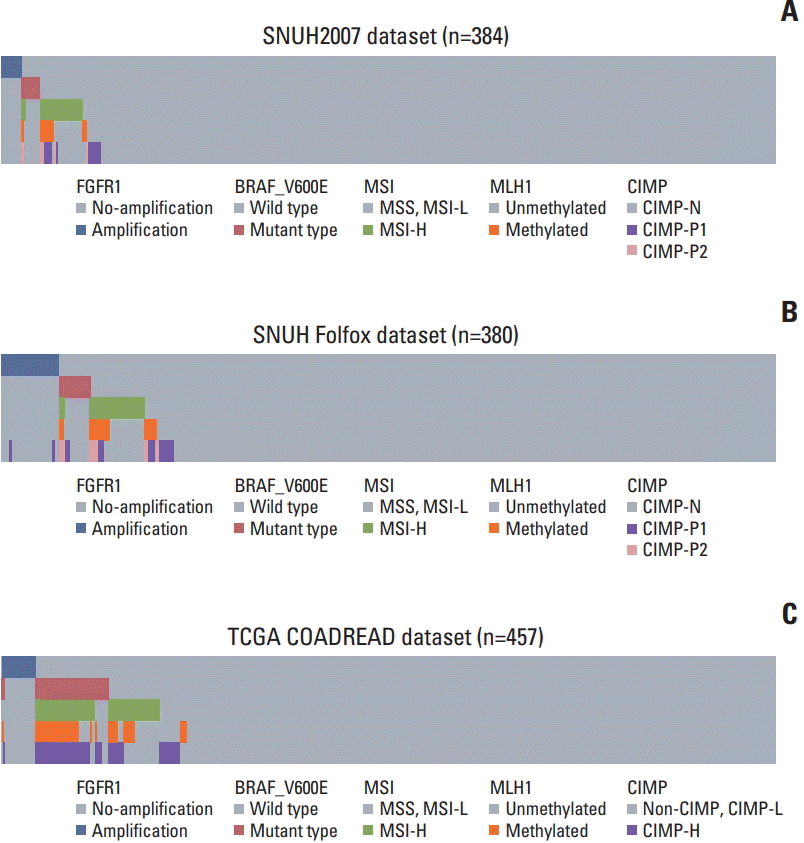
Fig. 2.
Kaplan-Meier survival curves. (A) 5-Year progression-free survival (PFS) according to fibroblast growth factor receptor 1 (FGFR1) copy number status in SNUH2007 dataset. (B) 5-Year disease-free survival (DFS) according to FGFR1 copy number status in SNUH Folfox dataset. (C) Overall survival (OS) according to FGFR1 copy number status in The Cancer Genome Atlas (TCGA) COADREAD dataset.
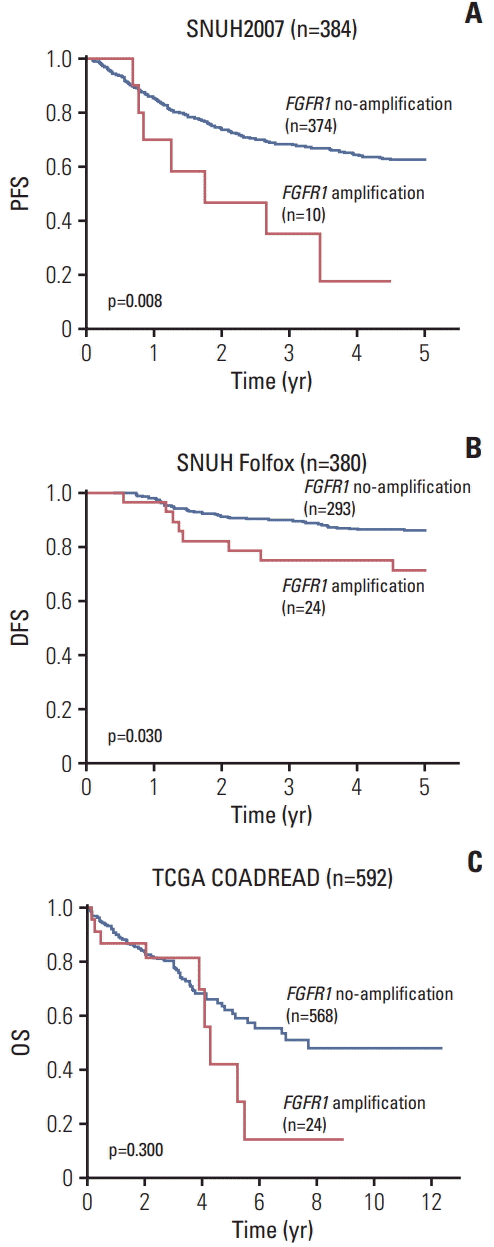
Fig. 3.
Cell viability assay using fibroblast growth factor receptor (FGFR) inhibitor, PD173074, in five colorectal cancer cell lines. (A) Copy number of FGFR1 measured by droplet digital polymerase chain reaction. (B) Western blot analysis of FGFR1 protein. (C) Dose-response curves to PD173074. (D) Summary of cell viability assay.
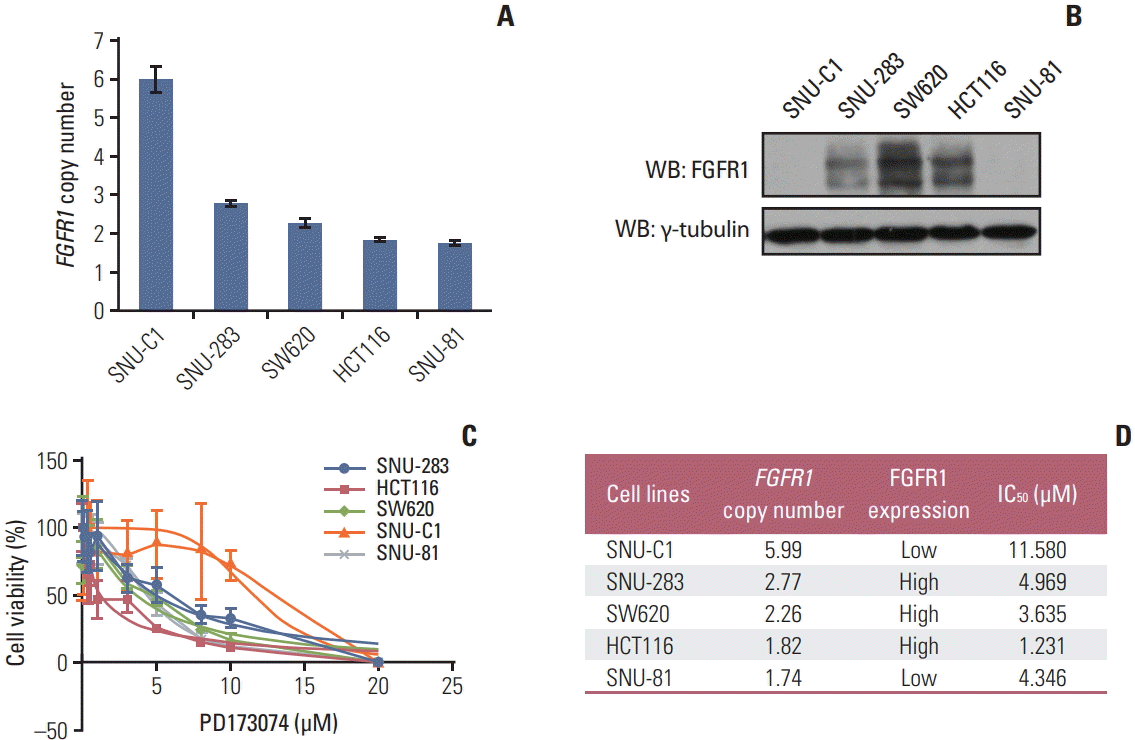
Table 1.
Clinicopathologic characteristics of colorectal cancers according to FGFR1 copy number and expression in SNUH-2007 dataset
| FGFR1 no-amplification (n=374, 97.4%) | FGFR1 amplification (n=10, 2.6%) | p-value | FGFR1 low-expression (n=332, 86.9%) | FGFR1 high-expression (n=50, 13.1%) | p-value | |
|---|---|---|---|---|---|---|
| Age (yr) | 63 (28-84) | 53 (29-75) | 0.048 | 63 (28-84) | 60 (37-83) | 0.271 |
| Sex | ||||||
| Male | 221 (59.1) | 4 (40.0) | 0.330a) | 205 (61.7) | 20 (40.0) | 0.004 |
| Female | 153 (40.9) | 6 (60.0) | 127 (38.3) | 30 (60.0) | ||
| Gross appearance | ||||||
| Fungating | 246 (65.8) | 7 (70.0) | > 0.999a) | 222 (66.9) | 30 (60.0) | 0.339 |
| Ulcerative | 128 (34.2) | 3 (30.0) | 110 (33.1) | 20 (40.0) | ||
| Tumor location | ||||||
| Proximal colon | 75 (20.0) | 2 (20.0) | 0.981 | 63 (19.0) | 14 (28.0) | 0.272 |
| Distal colon | 139 (37.2) | 4 (40.0) | 128 (38.5) | 15 (30.0) | ||
| Rectum | 160 (42.8) | 4 (40.0) | 141 (42.5) | 21 (42.0) | ||
| T category | ||||||
| T1 | 19 (5.1) | 0 | 0.129b) | 12 (3.6) | 6 (12.0) | 0.835b) |
| T2 | 51 (13.7) | 1 (10.0) | 49 (14.8) | 3 (6.0) | ||
| T3 | 259 (69.4) | 6 (60.0) | 229 (69.2) | 35 (70.0) | ||
| T4 | 44 (11.8) | 3 (30.0) | 41 (12.4) | 6 (12.0) | ||
| N category | ||||||
| N0 | 189 (50.5) | 3 (30.0) | 0.245b) | 173 (52.1) | 18 (36.0) | 0.073b) |
| N1 | 102 (27.3) | 4 (40.0) | 86 (25.9) | 19 (38.0) | ||
| N2 | 83 (22.2) | 3 (30.0) | 73 (22.0) | 13 (26.0) | ||
| M category | 0.407b) | |||||
| M0 | 307 (82.1) | 8 (80.0) | 0.867b) | 275 (82.8) | 39 (78.0) | |
| M1 | 67 (17.9) | 2 (20.0) | 57 (17.2) | 11 (22.0) | ||
| Stage | 0.072b) | |||||
| I | 57 (15.2) | 1 (10.0) | 0.377b) | 50 (15.1) | 7 (14.0) | |
| II | 123 (32.9) | 2 (20.0) | 116 (35.0) | 9 (18.0) | ||
| III | 127 (34.0) | 5 (50.0) | 109 (32.8) | 23 (46.0) | ||
| IV | 67 (17.9) | 2 (20.0) | 57 (17.2) | 11 (22.0) | ||
| Grade | ||||||
| Well differentiated | 10 (2.7) | 0 | 0.919b) | 10 (3.0) | 0 | 0.713b) |
| Moderately differentiated | 351 (93.8) | 10 (100) | 310 (93.4) | 49 (98.0) | ||
| Poorly differentiated | 13 (3.5) | 0 | 12 (3.6) | 1 (2.0) | ||
| KRAS mutation | ||||||
| Wild type | 271 (72.5) | 8 (80.0) | 0.734a) | 248 (74.7) | 30 (60.0) | 0.030 |
| Mutant type | 103 (27.5) | 2 (20.0) | 84 (25.3) | 20 (40.0) | ||
| BRAF mutation | 380 | 378 | ||||
| Wild type | 361 (97.6) | 10 (100) | > 0.999a) | 321 (97.9) | 48 (96.0) | 0.339a) |
| Mutant type | 9 (2.4) | 0 | 7 (2.1) | 2 (4.0) | ||
| MSI | ||||||
| MSS | 320 (85.6) | 9 (90.0) | 0.728 | 286 (86.1) | 42 (84.0) | 0.765 |
| MSI-low | 32 (8.6) | 1 (10.0) | 28 (8.4) | 4 (8.0) | ||
| MSI-high | 22 (5.9) | 0 | 18 (5.4) | 4 (8.0) | ||
| CIMP | ||||||
| CIMP-N | 358 (95.7) | 10 (100) | 0.800 | 321 (96.7) | 45 (90.0) | 0.070 |
| CIMP-P1 | 11 (2.9) | 0 | 8 (2.4) | 3 (6.0) | ||
| CIMP-P2 | 5 (1.4) | 0 | 3 (0.9) | 2 (4.0) |
Table 2.
Multivariate Cox model for 5-year disease-free survival in the SNUH Folfox dataset (n=380)




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download