1. Lee DS, Yoon WJ, Cho ES, Kim HJ, Gronostajski RM, Cho MI, Park JC. 2011; Crosstalk between nuclear factor I-C and transforming growth factor-β1 signaling regulates odontoblast differentiation and homeostasis. PLoS One. 6:e29160. DOI:
10.1371/journal.pone.0029160. PMID:
22195013. PMCID:
PMC3241690.

2. Ruch JV. 1998; Odontoblast commitment and differentiation. Biochem Cell Biol. 76:923–938. DOI:
10.1139/o99-008. PMID:
10392706.

3. Tummers M, Thesleff I. 2009; The importance of signal pathway modulation in all aspects of tooth development. J Exp Zool B Mol Dev Evol. 312B:309–319. DOI:
10.1002/jez.b.21280. PMID:
19156667.

4. Mina M, Kollar EJ. 1987; The induction of odontogenesis in non-dental mesenchyme combined with early murine mandibular arch epithelium. Arch Oral Biol. 32:123–127. DOI:
10.1016/0003-9969(87)90055-0. PMID:
3478009.

5. Sun Q, Liu H, Lin H, Yuan G, Zhang L, Chen Z. 2013; MicroRNA-338-3p promotes differentiation of mDPC6T into odontoblast-like cells by targeting Runx2. Mol Cell Biochem. 377:143–149. DOI:
10.1007/s11010-013-1580-3. PMID:
23380982.

6. Park MG, Kim JS, Park SY, Lee SA, Kim HJ, Kim CS, Kim JS, Chun HS, Park JC, Kim DK. 2014; MicroRNA-27 promotes the differentiation of odontoblastic cell by targeting APC and activating Wnt/β-catenin signaling. Gene. 538:266–272. DOI:
10.1016/j.gene.2014.01.045. PMID:
24487055.

7. Telles PD, Hanks CT, Machado MA, Nör JE. 2003; Lipoteichoic acid up-regulates VEGF expression in macrophages and pulp cells. J Dent Res. 82:466–470. DOI:
10.1177/154405910308200612. PMID:
12766200.

8. Ritchie HH, Ritchie DG, Wang LH. 1998; Six decades of dentinogenesis research. Historical and prospective views on phosphophoryn and dentin sialoprotein. Eur J Oral Sci. 106 Suppl 1:211–220. DOI:
10.1111/j.1600-0722.1998.tb02178.x. PMID:
9541228.
9. Tziafas D, Kolokuris I. 1990; Inductive influences of demineralized dentin and bone matrix on pulp cells: an approach of secondary dentinogenesis. J Dent Res. 69:75–81. DOI:
10.1177/00220345900690011301. PMID:
2303600.

10. Tziafas D, Papadimitriou S. 1998; Role of exogenous TGF-beta in induction of reparative dentinogenesis in vivo. Eur J Oral Sci. 106 Suppl 1:192–196. DOI:
10.1111/j.1600-0722.1998.tb02175.x. PMID:
9541225.
11. Narayanan K, inivas R Sr, Ramachandran A, Hao J, Quinn B, George A. 2001; Differentiation of embryonic mesenchymal cells to odontoblast-like cells by overexpression of dentin matrix protein 1. Proc Natl Acad Sci U S A. 98:4516–4521. DOI:
10.1073/pnas.081075198. PMID:
11287660. PMCID:
PMC31866.

12. eenath T Sr, Thyagarajan T, Hall B, Longenecker G, D'Souza R, Hong S, Wright JT, MacDougall M, Sauk J, Kulkarni AB. 2003; Dentin sialophosphoprotein knockout mouse teeth display widened predentin zone and develop defective dentin mineralization similar to human dentinogenesis imperfecta type III. J Biol Chem. 278:24874–24880. DOI:
10.1074/jbc.M303908200. PMID:
12721295.
13. About I, Bottero MJ, de Denato P, Camps J, Franquin JC, Mitsiadis TA. 2000; Human dentin production in vitro. Exp Cell Res. 258:33–41. DOI:
10.1006/excr.2000.4909. PMID:
10912785.

14. Revollo JR, Grimm AA, Imai S. 2004; The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 279:50754–50763. DOI:
10.1074/jbc.M408388200. PMID:
15381699.

15. Li Y, He X, Li Y, He J, Anderstam B, Andersson G, Lindgren U. 2011; Nicotinamide phosphoribosyltransferase (Nampt) affects the lineage fate determination of mesenchymal stem cells: a possible cause for reduced osteogenesis and increased adipogenesis in older individuals. J Bone Miner Res. 26:2656–2664. DOI:
10.1002/jbmr.480. PMID:
21812028.

16. Ling M, Huang P, Islam S, Heruth DP, Li X, Zhang LQ, Li DY, Hu Z, Ye SQ. 2017; Epigenetic regulation of Runx2 transcription and osteoblast differentiation by nicotinamide phosphoribosyltransferase. Cell Biosci. 7:27. DOI:
10.1186/s13578-017-0154-6. PMID:
28546856. PMCID:
PMC5442704.

18. Hanks CT, Sun ZL, Fang DN, Edwards CA, Wataha JC, Ritchie HH, Butler WT. 1998; Cloned 3T6 cell line from CD-1 mouse fetal molar dental papillae. Connect Tissue Res. 37:233–249. DOI:
10.3109/03008209809002442. PMID:
9862224.

19. Sun ZL, Fang DN, Wu XY, Ritchie HH, Bègue-Kirn C, Wataha JC, Hanks CT, Butler WT. 1998; Expression of dentin sialoprotein (DSP) and other molecular determinants by a new cell line from dental papillae, MDPC-23. Connect Tissue Res. 37:251–261. DOI:
10.3109/03008209809002443. PMID:
9862225.

20. Bosshardt DD, Nanci A. 2004; Hertwig's epithelial root sheath, enamel matrix proteins, and initiation of cementogenesis in porcine teeth. J Clin Periodontol. 31:184–192. DOI:
10.1111/j.0303-6979.2004.00473.x. PMID:
15016022.

21. Bègue-Kirn C, Smith AJ, Loriot M, Kupferle C, Ruch JV, Lesot H. 1994; Comparative analysis of TGF beta s, BMPs, IGF1, msxs, fibronectin, osteonectin and bone sialoprotein gene expression during normal and in vitro-induced odontoblast differentiation. Int J Dev Biol. 38:405–420. PMID:
7848824.
22. Xie H, Tang SY, Luo XH, Huang J, Cui RR, Yuan LQ, Zhou HD, Wu XP, Liao EY. 2007; Insulin-like effects of visfatin on human osteoblasts. Calcif Tissue Int. 80:201–210. DOI:
10.1007/s00223-006-0155-7. PMID:
17340225.

23. Gong Q, Wang R, Jiang H, Lin Z, Ling J. 2012; Alteration of microRNA expression of human dental pulp cells during odontogenic differentiation. J Endod. 38:1348–1354. DOI:
10.1016/j.joen.2012.06.016. PMID:
22980176.

24. Ok CY, Park S, Jang HO, Takata T, Lee OH, Bae MK, Bae SK. 2021; FK866 protects human dental pulp cells against oxidative stress-induced cellular senescence. Antioxidants (Basel). 10:271. DOI:
10.3390/antiox10020271. PMID:
33578781. PMCID:
PMC7916510.

25. Travelli C, Aprile S, Rahimian R, Grolla AA, Rogati F, Bertolotti M, Malagnino F, di Paola R, Impellizzeri D, Fusco R, Mercalli V, Massarotti A, Stortini G, Terrazzino S, Del Grosso E, Fakhfouri G, Troiani MP, Alisi MA, Grosa G, Sorba G, et al. 2017; Identification of novel triazole-based nicotinamide phosphoribosyltransferase (NAMPT) inhibitors endowed with antiproliferative and antiinflammatory activity. J Med Chem. 60:1768–1792. DOI:
10.1021/acs.jmedchem.6b01392. PMID:
28165742.

26. Li S, Kong H, Yao N, Yu Q, Wang P, Lin Y, Wang J, Kuang R, Zhao X, Xu J, Zhu Q, Ni L. 2011; The role of runt-related transcription factor 2 (Runx2) in the late stage of odontoblast differentiation and dentin formation. Biochem Biophys Res Commun. 410:698–704. DOI:
10.1016/j.bbrc.2011.06.065. PMID:
21703228.

27. Yamashiro T, Aberg T, Levanon D, Groner Y, Thesleff I. 2002; Expression of Runx1, -2 and -3 during tooth, palate and craniofacial bone development. Gene Expr Patterns. 2:109–112. DOI:
10.1016/S0925-4773(02)00298-8. PMID:
12617847.

29. Wen Q, Jing J, Han X, Feng J, Yuan Y, Ma Y, Chen S, Ho TV, Chai Y. 2020; Runx2 regulates mouse tooth root development via activation of WNT inhibitor NOTUM. J Bone Miner Res. 35:2252–2264. DOI:
10.1002/jbmr.4120. PMID:
32569388. PMCID:
PMC7689689.

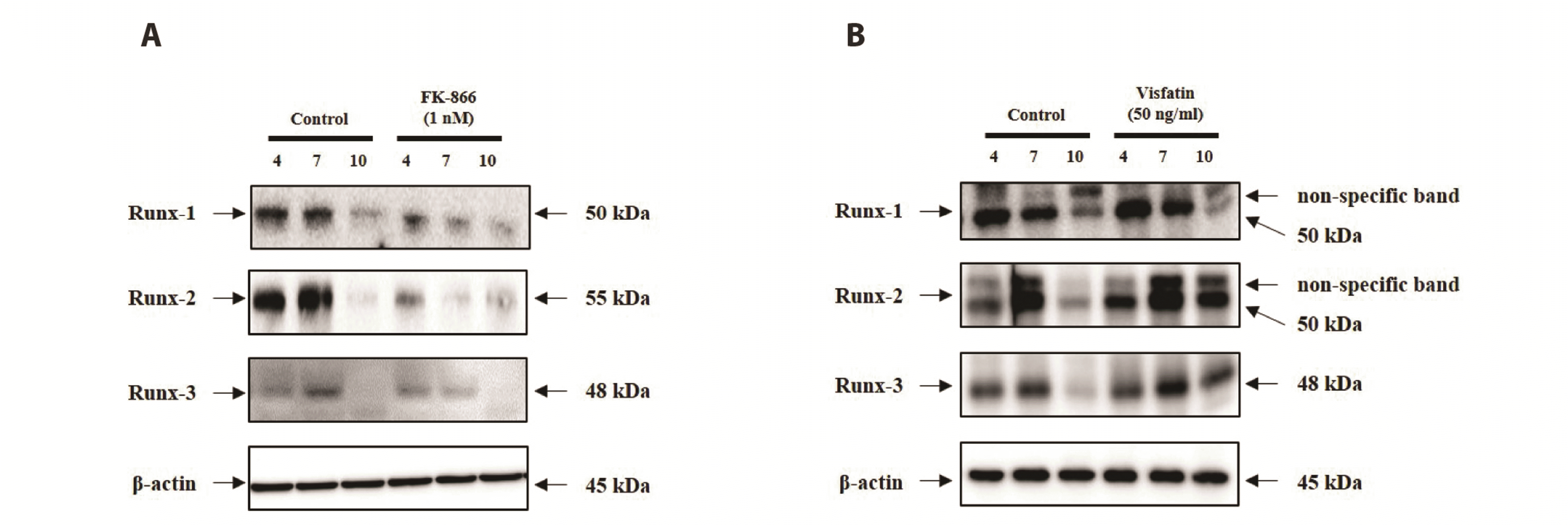




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download