This article has been
cited by other articles in ScienceCentral.
Abstract
An outbreak of fatal humidifier disinfectant lung injuries (HDLI) occurred in Korea. Human studies on mechanisms underlying HDLI have yet to be conducted. This study aimed to investigate methylation changes and their potential role in HDLI after exposure to HDs containing polyhexamethylene guanidine-phosphate. DNA methylation analysis was performed in blood samples from 10 children with HDLI and 10 healthy children using Infinium Human MethylationEPIC BeadChip. Transcriptome analysis was performed using lung tissues from 5 children with HDLI and 5 controls. Compared to healthy controls, 92 hypo-methylated and 79 hyper-methylated CpG sites were identified in children with HDLI at the statistical significance level of |Δβ|>0.2 and p<0.05. NOTCH1 was identified as a candidate network hub gene in cases. NOTCH1 transcripts significantly increased in lung tissues from HDLI cases compared to unexposed controls (p=0.05). NOTCH1 may play an important role in pulmonary fibrosis of HDLI.
Keywords: Humidifier disinfectant, pulmonary fibrosis, NOTCH1, methylation, polyhexamethylene guanidine
An outbreak of fatal lung injuries occurred in Korea between early 2000 and 2011, characterized by rapidly progressing respiratory failure with lung fibrosis, extensive air leak syndrome in many cases, a lack of responsiveness to any treatment, and high mortality rate.
1,
2,
3,
4,
5 This fatal interstitial lung disease (ILD) was distinct from previously identified ILDs in terms of clinical course as well as radiologic and pathologic findings; therefore, it was considered to be idiopathic.
1,
2 Toxic chemicals, including polyhexamethylene guanidine (PHMG), in humidifier disinfectants (HDs) were subsequently identified as the cause.
1,
2 The unique features of this fatal lung disease raised questions regarding the distinct mechanisms underlying the disorder.
6 However, there has been no report on the mechanisms underlying HD-associated lung injuries (HDLI) in humans. As altered DNA methylation is associated with development of idiopathic pulmonary fibrosis,
6,
7 we investigated whether DNA methylation plays a role in HDLI using human samples.
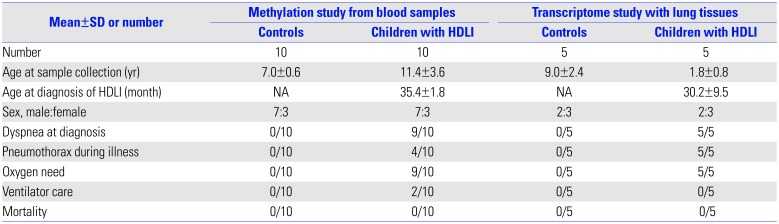
Blood samples from 10 children with HDLI and 10 healthy control children with no exposure to HDs were used to analyze methylation profiles. Clinical characteristics of the study population are summarized in
Table 1. The mean age at diagnosis of HDLI was 35.4 months (range, 12–81 months) and blood samples for methylation analysis were obtained at a mean age of 11.4 years (range, 7–15 years). Male-to-female rate was 7:3. None of the children in sex-matched control group had any respiratory diseases and their mean age was 7 years. DNA extracted from the peripheral blood mononuclear cells of each subject was analyzed using Infinium Human MethylationEPIC BeadChip (Illumina, San Diego, CA, USA). For quality check (QC) of the methylation data, beta-mixture quantile normalization, and Pearson's correlation (range: −1≤r≤1) for reproducibility between samples were performed. For QC of the transcriptome data, all data were normalized with the robust multi-average method implemented in in Affymetrix® Power Tools (Thermo Fisher Scientific, Waltham, MA, USA). Statistical significance for differentially methylated CpG sites was set at |Δβ|>0.2 and
p<0.05 using a t-test. Ingenuity® Pathway Analysis (IPA, Ingenuity Systems, Redwood City, CA, USA) was used to represent the functional networks of genes containing differentially methylated CpG sites. Transcriptome analysis was performed using lung tissues from five pediatric patients with HDLI and five control children. Lung tissue was obtained from children with no abnormal lung lesions from Bio-Resource Center at Asan Medical Center to form a control group. The Institutional Review Board of Asan Medical Center reviewed and approved the study protocol (IRB No. 2016-0885).
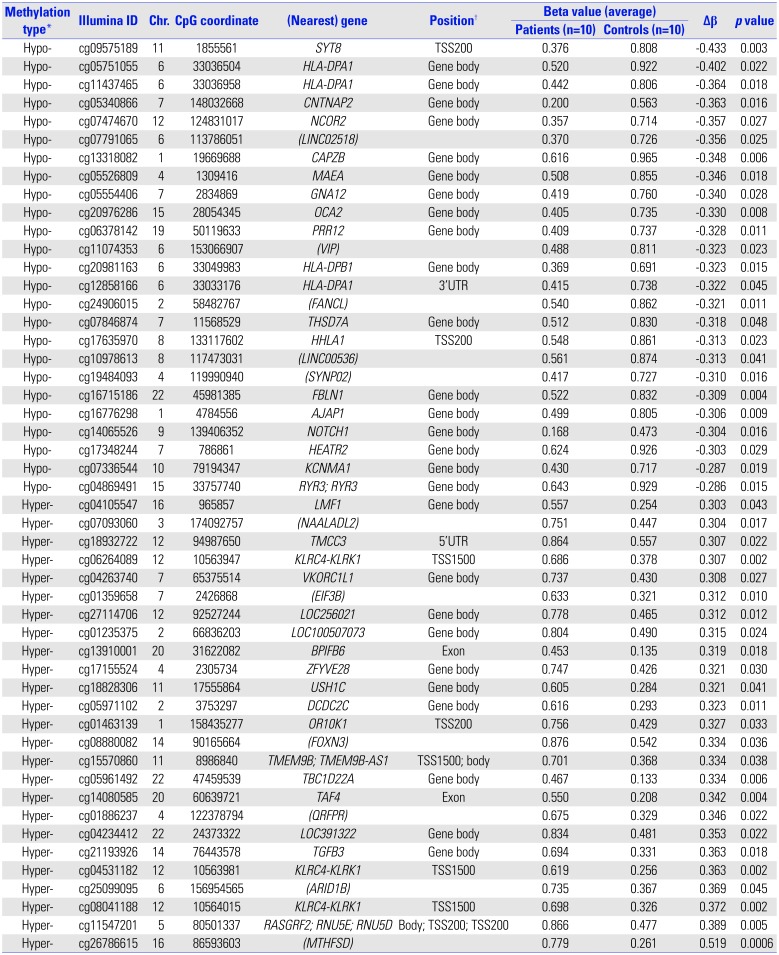
A total of 171 CpG loci (79 hypermethylated, 92 hypomethylated) showed significantly differential methylation patterns in children with HDLI compared to the controls (
Fig. 1A), with a distinctive clustering observed between the two groups (
Fig. 1B) (
Table 2). The top 25 hypomethylated and 25 hypermethylated CpG loci are listed in
Table 2. SYT8 cg09575189 showed the highest hypomethylation level (|Δβ|=0.433,
p=0.003), whereas cg26786615 (chr16: 86593603) had the highest hypermethylation level (|Δβ|=0.519,
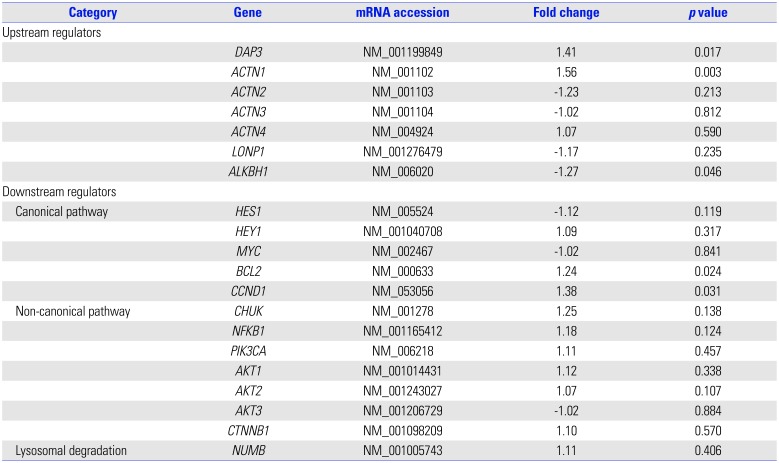
p=0.0006). However, there are a few functional studies of these two genes and no reports in existing literature that provide any clues to the associations between them and fibrosis and/or lung diseases. Potential upstream and downstream regulators of NOTCH1 based on IPA network analysis and its signaling (
https://www.rndsystems.com/pathwyas/notch-signaling-pathway) are described in
Table 3.
NOTCH1 cg14065526 showed a significant degree of hypomethylation (|Δβ|=0.304,
p=0.016). In further network analysis of the genes containing differently methylated CpG sites, “cancer, organismal injury and abnormalities, reproductive system disease (score=41)” was identified as the top network for HDLI, indicating
NOTCH1 as a hub gene (
Fig. 1C). The cg14065526 (chr9: 139406352) of
NOTCH1 showed a significantly hypomethylated level (|Δβ|=0.304,
p=0.016).
NOTCH1 transcripts from lung tissues were significantly elevated in HDLI cases compared to unexposed controls (
p=0.05, each group n=5) (
Fig. 1D).
Our present findings from methylation and transcriptome analysis of human blood and lung tissues have identified that NOTCH1 is involved in the pathogenesis of HDLI. This is the first study to investigate DNA methylation changes and network analyses combined with transcriptomics in pediatric patients with HDLI, which may partially explain the underlying mechanisms of HDLI.
Although
NOTCH1 may be common to the mechanisms of other types of ILDs,
8 the results of our current analysis suggest that it also plays a central role in the mechanism of HDLI. Notch1 is involved in angiogenesis, abnormal remodeling of vessels, and mucus hypersecretion, and thereby is associated with pathogenesis of diverse lung diseases.
9 The apoptosis of bronchial epithelial cells following exposure to toxic chemicals affects the clearance of apoptotic debris combined with lung fibrosis.
10 The overexpression of
NOTCH1, which is related to its gene hypomethylation, as shown in this study, promotes the differentiation of myofibroblasts, which is a critical step in pulmonary fibrosis.
3 NOTCH1 has been identified to be involved in bleomycin-induced lung diseases and paraquat poisoning, for which the main mechanism is pulmonary fibrosis.
11,
12 The results of previous reports and our present findings provide strong evidence for the involvement of
NOTCH1 in the pathogenesis of fatal fibrotic lung diseases and give new insights into the possible mechanisms of lung injuries caused by inhalation of unidentified but harmful chemicals that are commonly used.
The inhalation of toxic chemicals damages the epithelial lining in the airway, initiating a series of processes including disruption of epithelial lining, alterations of diverse mediators and chemokine levels, and induction of epithelium-to-mesenchymal transition (EMT).
13 NOTCH1 regulates EMT through various signaling factors, such as TGF-β, NF-κB, and β-catenin.
10 It has been reported that exposure to PHMG phosphate can induce EMT in a dose-dependent manner.
14 A previous study identified that PHMG could induce EMT through the Akt/Notch signaling pathway.
15 This prior evidence, in combination with our current data, further supports the notion that
NOTCH1 plays a role in the pathogenesis of HDLI via EMT following exposure to HDs that contain PHMG.
Our study had some limitations, including its small sample size. However, the results of the current study are significant in that HDLI is an exceptional disease, and the acquisition of blood and lung tissue in our patients was not easy. In our present cohort, there were time lags with a mean of 9 years between diagnosis of HDLI and blood sampling. The methylation patterns in the blood obtained after a time lag of 9 years may have been affected by diverse factors.
16 A previous study showed that less than 30% of individuals showed methylation changes in epigenome- wide DNA methylation analysis on average 11 years apart, even with intra-individual variations.
16 We could not perform methylation analysis in human lung tissues in the current study, as these samples were not available. In spite of the limitations, methylation changes observed in the present study could be helpful to elucidate the mechanisms underlying HDLI with stable disease state.
In conclusion, we have identified NOTCH1 pathways as one of the possible main fibrogenetic mechanisms of HDLI in children following exposure to PHMG phosphate. Further identification and elucidation of the mechanisms underlying this fatal lung disease are essential for the future development of therapeutics and prevention of lung diseases after exposure to harmful domestic chemicals.
Fig. 1
Results of methylation, network, and NOTCH1 expression analysis in pediatric HDLI cases. (A) Volcano plot of differentially methylated CpG sites. (B) Heatmap of differentially methylated CpG sites between children with humidifier disinfectant associated lung injuries and unexposed healthy controls. Differentially methylated CpG loci indicated by asterisk. (C) The top network of differentially methylated CpG sites was found to be “cancer, organismal injury and abnormalities, reproductive system disease” and was derived from genes containing hyper-/hypo-methylated CpG sites associated with HDLI. (D) The transcriptional expression of NOTCH1 between HDLI cases and the control group (p=0.05, t-test, nonparametric methods were applied, and no correction for multiple testing was done due to the small sample size of each group, n=5 for each group). HDLI, humidifier disinfectant lung injuries.
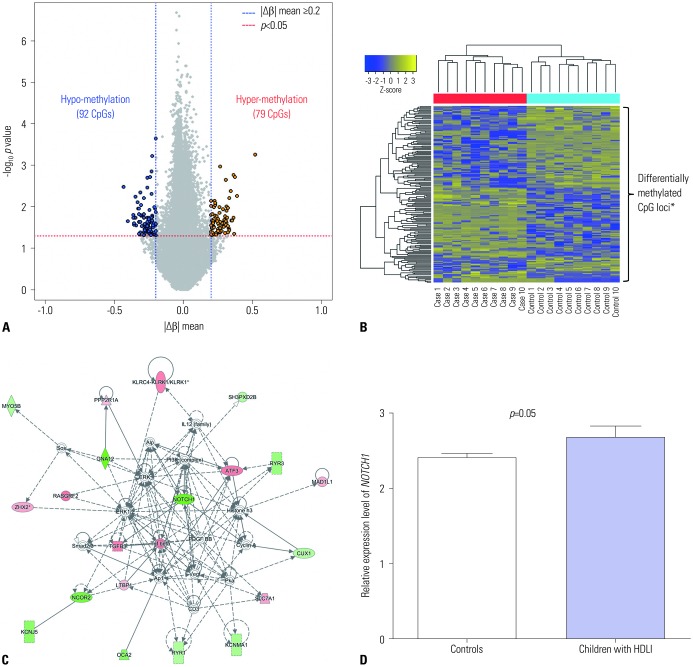
Table 1
Clinical Characteristics of the Study Population
|
Mean±SD or number |
Methylation study from blood samples |
Transcriptome study with lung tissues |
|
Controls |
Children with HDLI |
Controls |
Children with HDLI |
|
Number |
10 |
10 |
5 |
5 |
|
Age at sample collection (yr) |
7.0±0.6 |
11.4±3.6 |
9.0±2.4 |
1.8±0.8 |
|
Age at diagnosis of HDLI (month) |
NA |
35.4±1.8 |
NA |
30.2±9.5 |
|
Sex, male:female |
7:3 |
7:3 |
2:3 |
2:3 |
|
Dyspnea at diagnosis |
0/10 |
9/10 |
0/5 |
5/5 |
|
Pneumothorax during illness |
0/10 |
4/10 |
0/5 |
5/5 |
|
Oxygen need |
0/10 |
9/10 |
0/5 |
5/5 |
|
Ventilator care |
0/10 |
2/10 |
0/5 |
0/5 |
|
Mortality |
0/10 |
0/10 |
0/5 |
0/5 |
Table 2
Top 25 Hypomethylated and Top 25 Hypermethylated Sites Showing Significantly Different Levels in Pediatric Patients with HDLI Compared to Unexposed Healthy Control Children
|
Methylation type*
|
Illumina ID |
Chr. |
CpG coordinate |
(Nearest) gene |
Position†
|
Beta value (average) |
Δβ |
p value |
|
Patients (n=10) |
Controls (n=10) |
|
Hypo- |
cg09575189 |
11 |
1855561 |
SYT8
|
TSS200 |
0.376 |
0.808 |
-0.433 |
0.003 |
|
Hypo- |
cg05751055 |
6 |
33036504 |
HLA-DPA1
|
Gene body |
0.520 |
0.922 |
-0.402 |
0.022 |
|
Hypo- |
cg11437465 |
6 |
33036958 |
HLA-DPA1
|
Gene body |
0.442 |
0.806 |
-0.364 |
0.018 |
|
Hypo- |
cg05340866 |
7 |
148032668 |
CNTNAP2
|
Gene body |
0.200 |
0.563 |
-0.363 |
0.016 |
|
Hypo- |
cg07474670 |
12 |
124831017 |
NCOR2
|
Gene body |
0.357 |
0.714 |
-0.357 |
0.027 |
|
Hypo- |
cg07791065 |
6 |
113786051 |
(LINC02518)
|
|
0.370 |
0.726 |
-0.356 |
0.025 |
|
Hypo- |
cg13318082 |
1 |
19669688 |
CAPZB
|
Gene body |
0.616 |
0.965 |
-0.348 |
0.006 |
|
Hypo- |
cg05526809 |
4 |
1309416 |
MAEA
|
Gene body |
0.508 |
0.855 |
-0.346 |
0.018 |
|
Hypo- |
cg05554406 |
7 |
2834869 |
GNA12
|
Gene body |
0.419 |
0.760 |
-0.340 |
0.028 |
|
Hypo- |
cg20976286 |
15 |
28054345 |
OCA2
|
Gene body |
0.405 |
0.735 |
-0.330 |
0.008 |
|
Hypo- |
cg06378142 |
19 |
50119633 |
PRR12
|
Gene body |
0.409 |
0.737 |
-0.328 |
0.011 |
|
Hypo- |
cg11074353 |
6 |
153066907 |
(VIP)
|
|
0.488 |
0.811 |
-0.323 |
0.023 |
|
Hypo- |
cg20981163 |
6 |
33049983 |
HLA-DPB1
|
Gene body |
0.369 |
0.691 |
-0.323 |
0.015 |
|
Hypo- |
cg12858166 |
6 |
33033176 |
HLA-DPA1
|
3′UTR |
0.415 |
0.738 |
-0.322 |
0.045 |
|
Hypo- |
cg24906015 |
2 |
58482767 |
(FANCL)
|
|
0.540 |
0.862 |
-0.321 |
0.011 |
|
Hypo- |
cg07846874 |
7 |
11568529 |
THSD7A
|
Gene body |
0.512 |
0.830 |
-0.318 |
0.048 |
|
Hypo- |
cg17635970 |
8 |
133117602 |
HHLA1
|
TSS200 |
0.548 |
0.861 |
-0.313 |
0.023 |
|
Hypo- |
cg10978613 |
8 |
117473031 |
(LINC00536)
|
|
0.561 |
0.874 |
-0.313 |
0.041 |
|
Hypo- |
cg19484093 |
4 |
119990940 |
(SYNP02)
|
|
0.417 |
0.727 |
-0.310 |
0.016 |
|
Hypo- |
cg16715186 |
22 |
45981385 |
FBLN1
|
Gene body |
0.522 |
0.832 |
-0.309 |
0.004 |
|
Hypo- |
cg16776298 |
1 |
4784556 |
AJAP1
|
Gene body |
0.499 |
0.805 |
-0.306 |
0.009 |
|
Hypo- |
cg14065526 |
9 |
139406352 |
NOTCH1
|
Gene body |
0.168 |
0.473 |
-0.304 |
0.016 |
|
Hypo- |
cg17348244 |
7 |
786861 |
HEATR2
|
Gene body |
0.624 |
0.926 |
-0.303 |
0.029 |
|
Hypo- |
cg07336544 |
10 |
79194347 |
KCNMA1
|
Gene body |
0.430 |
0.717 |
-0.287 |
0.019 |
|
Hypo- |
cg04869491 |
15 |
33757740 |
RYR3; RYR3
|
Gene body |
0.643 |
0.929 |
-0.286 |
0.015 |
|
Hyper- |
cg04105547 |
16 |
965857 |
LMF1
|
Gene body |
0.557 |
0.254 |
0.303 |
0.043 |
|
Hyper- |
cg07093060 |
3 |
174092757 |
(NAALADL2)
|
|
0.751 |
0.447 |
0.304 |
0.017 |
|
Hyper- |
cg18932722 |
12 |
94987650 |
TMCC3
|
5′UTR |
0.864 |
0.557 |
0.307 |
0.022 |
|
Hyper- |
cg06264089 |
12 |
10563947 |
KLRC4-KLRK1
|
TSS1500 |
0.686 |
0.378 |
0.307 |
0.002 |
|
Hyper- |
cg04263740 |
7 |
65375514 |
VKORC1L1
|
Gene body |
0.737 |
0.430 |
0.308 |
0.027 |
|
Hyper- |
cg01359658 |
7 |
2426868 |
(EIF3B)
|
|
0.633 |
0.321 |
0.312 |
0.010 |
|
Hyper- |
cg27114706 |
12 |
92527244 |
LOC256021
|
Gene body |
0.778 |
0.465 |
0.312 |
0.012 |
|
Hyper- |
cg01235375 |
2 |
66836203 |
LOC100507073
|
Gene body |
0.804 |
0.490 |
0.315 |
0.024 |
|
Hyper- |
cg13910001 |
20 |
31622082 |
BPIFB6
|
Exon |
0.453 |
0.135 |
0.319 |
0.018 |
|
Hyper- |
cg17155524 |
4 |
2305734 |
ZFYVE28
|
Gene body |
0.747 |
0.426 |
0.321 |
0.030 |
|
Hyper- |
cg18828306 |
11 |
17555864 |
USH1C
|
Gene body |
0.605 |
0.284 |
0.321 |
0.041 |
|
Hyper- |
cg05971102 |
2 |
3753297 |
DCDC2C
|
Gene body |
0.616 |
0.293 |
0.323 |
0.011 |
|
Hyper- |
cg01463139 |
1 |
158435277 |
OR10K1
|
TSS200 |
0.756 |
0.429 |
0.327 |
0.033 |
|
Hyper- |
cg08880082 |
14 |
90165664 |
(FOXN3)
|
|
0.876 |
0.542 |
0.334 |
0.036 |
|
Hyper- |
cg15570860 |
11 |
8986840 |
TMEM9B; TMEM9B-AS1
|
TSS1500; body |
0.701 |
0.368 |
0.334 |
0.038 |
|
Hyper- |
cg05961492 |
22 |
47459539 |
TBC1D22A
|
Gene body |
0.467 |
0.133 |
0.334 |
0.006 |
|
Hyper- |
cg14080585 |
20 |
60639721 |
TAF4
|
Exon |
0.550 |
0.208 |
0.342 |
0.004 |
|
Hyper- |
cg01886237 |
4 |
122378794 |
(QRFPR)
|
|
0.675 |
0.329 |
0.346 |
0.022 |
|
Hyper- |
cg04234412 |
22 |
24373322 |
LOC391322
|
Gene body |
0.834 |
0.481 |
0.353 |
0.022 |
|
Hyper- |
cg21193926 |
14 |
76443578 |
TGFB3
|
Gene body |
0.694 |
0.331 |
0.363 |
0.018 |
|
Hyper- |
cg04531182 |
12 |
10563981 |
KLRC4-KLRK1
|
TSS1500 |
0.619 |
0.256 |
0.363 |
0.002 |
|
Hyper- |
cg25099095 |
6 |
156954565 |
(ARID1B)
|
|
0.735 |
0.367 |
0.369 |
0.045 |
|
Hyper- |
cg08041188 |
12 |
10564015 |
KLRC4-KLRK1
|
TSS1500 |
0.698 |
0.326 |
0.372 |
0.002 |
|
Hyper- |
cg11547201 |
5 |
80501337 |
RASGRF2; RNU5E; RNU5D
|
Body; TSS200; TSS200 |
0.866 |
0.477 |
0.389 |
0.005 |
|
Hyper- |
cg26786615 |
16 |
86593603 |
(MTHFSD)
|
|
0.779 |
0.261 |
0.519 |
0.0006 |
Table 3
Gene Expression of Potential Upstream and Downstream Regulators of NOTCH1 in Formalin-Fixed, Paraffin-Embedded Lung Tissue Specimens from Children with HDLI and the Control Group
|
Category |
Gene |
mRNA accession |
Fold change |
p value |
|
Upstream regulators |
|
|
|
|
|
DAP3
|
NM_001199849 |
1.41 |
0.017 |
|
ACTN1
|
NM_001102 |
1.56 |
0.003 |
|
ACTN2
|
NM_001103 |
-1.23 |
0.213 |
|
ACTN3
|
NM_001104 |
-1.02 |
0.812 |
|
ACTN4
|
NM_004924 |
1.07 |
0.590 |
|
LONP1
|
NM_001276479 |
-1.17 |
0.235 |
|
ALKBH1
|
NM_006020 |
-1.27 |
0.046 |
|
Downstream regulators |
|
|
|
|
|
Canonical pathway |
HES1
|
NM_005524 |
-1.12 |
0.119 |
|
HEY1
|
NM_001040708 |
1.09 |
0.317 |
|
MYC
|
NM_002467 |
-1.02 |
0.841 |
|
BCL2
|
NM_000633 |
1.24 |
0.024 |
|
CCND1
|
NM_053056 |
1.38 |
0.031 |
|
Non-canonical pathway |
CHUK
|
NM_001278 |
1.25 |
0.138 |
|
NFKB1
|
NM_001165412 |
1.18 |
0.124 |
|
PIK3CA
|
NM_006218 |
1.11 |
0.457 |
|
AKT1
|
NM_001014431 |
1.12 |
0.338 |
|
AKT2
|
NM_001243027 |
1.07 |
0.107 |
|
AKT3
|
NM_001206729 |
-1.02 |
0.884 |
|
CTNNB1
|
NM_001098209 |
1.10 |
0.570 |
|
Lysosomal degradation |
NUMB
|
NM_001005743 |
1.11 |
0.406 |








 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download