Abstract
Various molecular targeted therapies and diagnostic modalities have been developed for the treatment of hepatocellular carcinoma (HCC); however, HCC still remains a difficult malignancy to cure. Recently, the focus has shifted to cancer metabolism for the diagnosis and treatment of various cancers, including HCC. In addition to conventional diagnostics, the measurement of enhanced tumor cell metabolism using F-18 fluorodeoxyglucose (18F-FDG) for increased glycolysis or C-11 acetate for fatty acid synthesis by positron emission tomography/computed tomography (PET/CT) is well established for clinical management of HCC. Unlike tumors displaying the Warburg effect, HCCs vary substantially in terms of 18F-FDG uptake, which considerably reduces the sensitivity for tumor detection. Accordingly, C-11 acetate has been proposed as a complementary radiotracer for detecting tumors that are not identified by 18F-FDG. In addition to HCC diagnosis, since the degree of 18F-FDG uptake converted to standardized uptake value (SUV) correlates well with tumor aggressiveness, 18F-FDG PET/CT scans can predict patient outcomes such as treatment response and survival with an inverse relationship between SUV and survival. The loss of tumor suppressor genes or activation of oncogenes plays an important role in promoting HCC development, and might be involved in the “metabolic reprogramming” of cancer cells. Mutations in various genes such as TERT, CTNNB1, TP53, and Axin1 are responsible for the development of HCC. Some microRNAs (miRNAs) involved in cancer metabolism are deregulated in HCC, indicating that the modulation of genes/miRNAs might affect HCC growth or metastasis. In this review, we will discuss cancer metabolism as a mechanism for treatment resistance, as well as an attractive potential therapeutic target in HCC.
Hepatocellular carcinoma (HCC) is the fifth leading cause of cancer mortality worldwide.12 The main cause of HCC is cirrhosis, which originates from infections caused by chronic hepatitis B virus, hepatitis C virus, or alcohol consumption.34 Patients with early-stage HCC are often asymptomatic, and it is usually detected at intermediate or advanced stages in which patients cannot be treated by curative hepatic resection or liver transplantation.5 In addition, although surgical treatment for early HCC has improved patient outcome, the risk of recurrence remains substantial and there is still no curative therapy for advanced HCC. Therefore, there is an increasing need for effective early diagnosis and development of novel therapeutics for HCC patients. Owing to its important role in metabolic reprogramming during carcinogenesis, cancer metabolism has gained popularity in the fields of cancer diagnosis and therapy. This review summarizes the current state of research related to cancer metabolism, to help identify potential new therapeutic targets for HCC.
Alpha-fetoprotein (AFP) was the first glycoprotein identified as a marker of HCC, and this protein is used to screen for this particular disease.6 However, approximately 30% of HCC patients maintain normal AFP levels, and some HCC patients also have relatively low levels of AFP.7 To overcome these problems, several imaging techniques are used to diagnose patients with suspected HCC, such as ultrasound, computed tomography (CT), magnetic resonance imaging (MRI), positron emission tomography/computed tomography (PET/CT), and angiography.
Of the imaging modalities, clinical efficacy of functional imaging based on cancer metabolism for the assessment of HCC has been actively investigated. F-18 fluorodeoxyglucose (18F-FDG), a surrogate for enhanced glucose metabolism, has been used widely for HCC. The uptake mechanism and biochemical pathway of 18F-FDG metabolism has been extensively studied in vitro and in vivo; transport through cell membrane via glucose transporter isoform 1 (GLUT1) and intracellular phosphorylation by hexokinase (HK) have been identified as key steps for subsequent accumulation in HCC.8910111213 Many studies have shown upregulation of GLUT1 in HCC, but not in non-tumor liver tissue.1415 Amann, et al.15 demonstrated a positive correlation between GLUT1 expression and Ki-67 labeling index in patients with HCC, suggesting that its expression is associated with advanced tumor stage and poor differentiation. In addition, poor survival has been reported in patients with high tumor GLUT1 expression, based on The Cancer Genome Atlas (TCGA) data set.16 A relationship between enhanced FDG uptake and dysregulation of epithelial-mesenchymal transition-related proteins was demonstrated in HCC through in vitro and patient tissue experiments.16 Representative of the underlying biological characteristics of tumor, 18F-FDG PET/CT images are predictive of tumor recurrence or survival after various treatments.17
Despite displaying increased glycolysis even with the presence of oxygen, the so-called Warburg effect, HCCs are notorious for exhibiting a wide spectrum of 18F-FDG uptake capabilities, considerably reducing the sensitivity of tumor detection. Alternatively, C-11 acetate has been proposed as a radiotracer for detecting tumors that are not identified based on 18F-FDG uptake (Figs. 1 and 2). Acetate is a source of acetyl-CoA, and it plays an essential role in regulating the activity and expression of proteins involved in regulation of intracellular biomass, lipogenesis, and acetylation.18 Acetate was shown to be utilized by tumors as an alternative nutrient under low cellular glucose uptake conditions, and C-11 acetate accumulation in tumors has been found to be associated with tumor progression.19 HCC has been reported to use acetate as a substrate for fatty acid biosynthesis through up-regulation of acetyl-CoA synthase and monocarboxylate transporter (MCT).20 Recent studies have indicated that MCT1 is a novel import system of acetate in non-glycolytic HCC tumors. Indeed, Fig. 3 shows various expressions of GLUT1 and MCT1 in HCC patients with different levels of 18F-FDG and 11C-acetate uptake. It was demonstrated that absorption of acetate by MCT1 promotes oxidative phosphorylation and lipid metabolism in non-glycolytic HCC tumors.21 Accordingly, combining 18F-FDG PET/CT with C-11 acetate PET/CT could be useful to provide relevant information on prognostic and molecular heterogeneity.
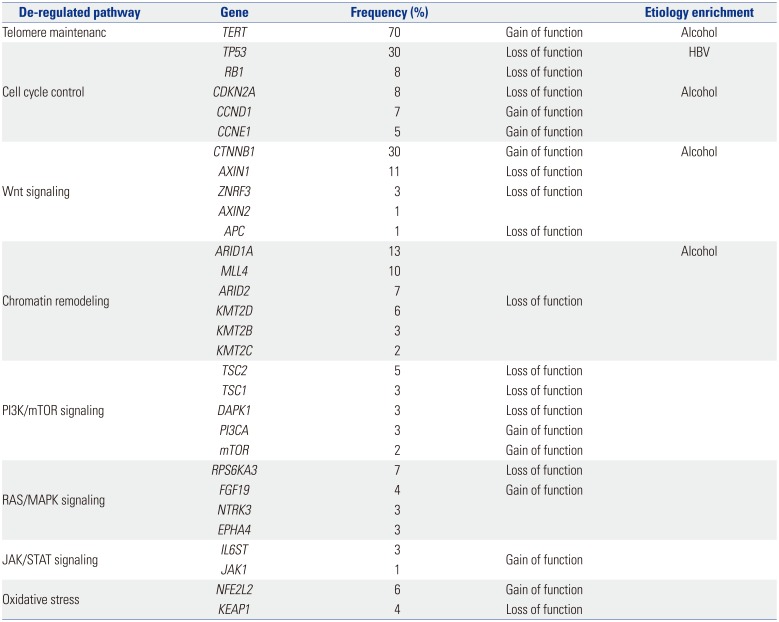
HCC is a heterogeneous disease, both clinically and from a molecular standpoint. Different risk factors such as hepatitis virus infection, aflatoxin exposure, or alcohol abuse are linked to specific pathways, and these can be strongly associated with certain types of HCC. Based on the results of HCC tumor sequencing, different driver genes and associated oncogenic pathways have been identified, based on the composition of tumor source.22232425 Therefore, heterogeneity should be investigated to determine the etiological cause and affected pathways of HCC. High levels of heterogeneity are clinically relevant, as they lead to inconsistent treatment outcomes. Recently, deep sequencing/next generation sequencing has provided new insights into the complex molecular pathogenesis of HCC, including the identification of novel oncogenic pathways and driver genes.252627282930313233 Aberrant telomerase reverse transcriptase (TERT) activation is the most common somatic alteration observed in HCC (~70%). In addition to TERT, CTNNB1, TP53, and Axin1 are mutated at high frequency in HCC. Table 1 summarizes the most relevant mutations in HCC.
Warburg effect occurs downstream of survival signaling pathways, which are altered by loss of tumor suppressor genes or activation of oncogenes such as c-Myc, Ras, Akt, TP53, and HIF-1α.3435363738
c-Myc was reported to result in mouse liver tumors with elevated glycolysis39. HIF-1α, a major transcription factor involved in hypoxic response of cancer cells,40 has been shown to play an important role in several cancers by promoting tumorigenesis, and might also be involved in the “metabolic reprogramming” of cancer cells.4142 This activates the transcription of genes encoding angiogenic cytokines and growth factors, such as VEGF and glycolytic enzymes including hexokinase1 (HK1), hexokinase2 (HK2), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and pyruvate kinase (PKM).434445 Moreover, HIF-1α enhances chemoresistance and radioresistance, whereas it suppresses differentiation and apoptosis in HCC.4647 As a result, elevated HIF-1α levels are associated with increased patient mortality and metastasis in various tumors, including HCC.484950
Certain gene mutations or loss-of-heterozygosity events alter metabolism in a HIF-1α-dependent manner. Although mutations in PTEN gene rarely occur in HCC, frequent loss of heterozygosity of PTEN allele has been identified (in 20–30% of HCC patients).5152 The loss of PTEN plays a critical role in HCC progression and patient outcome by increasing HIF-1α synthesis and stability.5354 The modulation of HIF-1α expression by epidermal growth factor/phosphatidylinositol 3-kinase (PI3K)/PTEN/AKT/FRAP pathway has implications in tumor angiogenesis.55 A few studies have shown that HIF-1 can be activated by Ras and membrane type-1 matrix metalloproteinases under normal oxygen conditions in cancer cells, providing new insight into regulation of cancer glycolysis beyond hypoxic condition.5657 Bufalin, a cardiotonic steroid, was shown to suppress tumor invasion and metastasis by targeting HIF-1α via PI3K/AKT/mTOR pathway, and thus has potential for HCC targeted therapy.58
Numerous microRNAs (miRNAs) have been shown to be associated with HCC. Six miRNAs have been consistently reported to be dysregulated in HCC, when compared to their expressions in non-tumorous tissue.596061 For example, miR-122 and miR-199a, which act as tumor suppressors by regulating the expression of cyclin G and components of PAK4/Raf/MEK/ERK pathway, are downregulated in this disease. Conversely, miR-21, miR-221, miR-222, and miR-224, which target various molecules including PTEN, SMAD4, CDKN1B, and CDKN1C, are upregulated in HCC. Some miRNAs that regulate cancer metabolism are also dysregulated in HCC. miR-34a plays a major role in regulation of cellular metabolism by targeting SIRT1, a key NAD-dependent deacetylating enzyme involved in a wide range of metabolic processes including lipid metabolism, glucose metabolism, and expression of other metabolic regulators.6263 This molecule inhibits cellular glycolysis by targeting HK1/2 and glucose-6-phosphate isomerase. miR-23a directly targets the key gluconeogenic enzyme glucose-6-phosphatase catalytic subunit (G6PC), and is significantly upregulated in primary human HCC.
Although sorafenib has limited efficacy, it is still the only standard treatment available for advanced HCC with portal vein invasion or extrahepatic spread.6465 However, other molecules are being developed for targeted therapy. Recent studies on metabolic regulation of cancer cell growth and metastasis have been actively performed. 2-deoxy-D-glucose (2-DG), a glucose analogue that is able to suppress glycolysis by competitively inhibiting HK2, has an effect on HCC growth.6667 The combination of conventional therapy and 2-DG has been reported to synergistically inhibit the proliferation of sorafenib-sensitive and sorafenib-resistant HCC cells.68 3-bromopyruvate (3-BP) directly inhibits HK2 activity and glycolysis pathway. In vitro and in vivo studies have demonstrated the anticancer effects of 3-BP on HCC, and consequently, this drug has been approved by the FDA.6970
Facilitative glucose transporters (GLUTs) have emerged as key factors that are required for increasing glucose uptake by cancer cells.1415 Therefore, small molecules targeting GLUT1 will inhibit cancer cell growth or metastasis by reducing glucose uptake. To increase GLUT1 targeting specificity, derivatives of GLUT1 inhibitor, such as fasentin7172 and WZB117,73 have been investigated. Regulation of hypoxia by molecules, including HIF-1, is an attractive potential therapeutic target for HCC as well as other cancers. The HIF-1α mRNA antagonist EZN-2968, a novel inhibitor of hypoxia-induced gene activation, is currently in Phase I trial for HCC patients.
Metabolic reprogramming is essential for angiogenesis, proliferation, invasion, and metastasis of cancer. It is also associated with de-differentiation, anti-apoptotic properties, and resistance to conventional chemotherapy and radiotherapy. In the future, cancer metabolism would represent an attractive potential therapeutic target. With their development, PET/CT scans combined with various metabolic radiotracers will offer clinical importance in selecting patients who would benefit from novel drugs targeting different pathways related to cancer metabolism
ACKNOWLEDGEMENTS
This research was supported by the National Research Foundation of Korea (Seoul, Korea; grant number NRF-2016R1E1A1A01943303, NRF-2015R1D1A1A01057737), and was supported by an Incheon National University Research Grant (Incheon, Korea) in 2016.
References
1. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010; 127:2893–2917. PMID: 21351269.

2. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015; 136:E359–E386. PMID: 25220842.

3. Velázquez RF, Rodríguez M, Navascués CA, Linares A, Pérez R, Sotorríos NG, et al. Prospective analysis of risk factors for hepatocellular carcinoma in patients with liver cirrhosis. Hepatology. 2003; 37:520–527. PMID: 12601348.

4. Fattovich G, Stroffolini T, Zagni I, Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004; 127(5 Suppl 1):S35–S50. PMID: 15508101.

5. Bruix J, Sherman M. American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update. Hepatology. 2011; 53:1020–1022. PMID: 21374666.

6. Song PP, Xia JF, Inagaki Y, Hasegawa K, Sakamoto Y, Kokudo N, et al. Controversies regarding and perspectives on clinical utility of biomarkers in hepatocellular carcinoma. World J Gastroenterol. 2016; 22:262–274. PMID: 26755875.

7. Cameron AM. Screening for viral hepatitis and hepatocellular cancer. Surg Clin North Am. 2015; 95:1013–1021. PMID: 26315520.

8. Phelps ME, Huang SC, Hoffman EJ, Selin C, Sokoloff L, Kuhl DE. Tomographic measurement of local cerebral glucose metabolic rate in humans with (F-18)2-fluoro-2-deoxy-D-glucose: validation of method. Ann Neurol. 1979; 6:371–388. PMID: 117743.

9. Gambhir SS, Czernin J, Schwimmer J, Silverman DH, Coleman RE, Phelps ME. A tabulated summary of the FDG PET literature. J Nucl Med. 2001; 42(5 Suppl):1S–93S. PMID: 11483694.
10. Stokkel MP, Draisma A, Pauwels EK. Positron emission tomography with 2-[18F]-fluoro-2-deoxy-D-glucose in oncology. Part IIIb: therapy response monitoring in colorectal and lung tumours, head and neck cancer, hepatocellular carcinoma and sarcoma. J Cancer Res Clin Oncol. 2001; 127:278–285. PMID: 11355142.
11. Iwata Y, Shiomi S, Sasaki N, Jomura H, Nishiguchi S, Seki S, et al. Clinical usefulness of positron emission tomography with fluorine-18-fluorodeoxyglucose in the diagnosis of liver tumors. Ann Nucl Med. 2000; 14:121–126. PMID: 10830530.

12. Khan MA, Combs CS, Brunt EM, Lowe VJ, Wolverson MK, Solomon H, et al. Positron emission tomography scanning in the evaluation of hepatocellular carcinoma. J Hepatol. 2000; 32:792–797. PMID: 10845666.

13. Böhm B, Voth M, Geoghegan J, Hellfritzsch H, Petrovich A, Scheele J, et al. Impact of positron emission tomography on strategy in liver resection for primary and secondary liver tumors. J Cancer Res Clin Oncol. 2004; 130:266–272. PMID: 14767761.
14. Medina RA, Owen GI. Glucose transporters: expression, regulation and cancer. Biol Res. 2002; 35:9–26. PMID: 12125211.

15. Amann T, Maegdefrau U, Hartmann A, Agaimy A, Marienhagen J, Weiss TS, et al. GLUT1 expression is increased in hepatocellular carcinoma and promotes tumorigenesis. Am J Pathol. 2009; 174:1544–1552. PMID: 19286567.

16. Lee M, Jeon JY, Neugent ML, Kim JW, Yun M. 18F-Fluorodeoxyglucose uptake on positron emission tomography/computed tomography is associated with metastasis and epithelial-mesenchymal transition in hepatocellular carcinoma. Clin Exp Metastasis. 2017; 34:251–260. PMID: 28429188.

17. Hwang SH, Lee JW, Cho HJ, Kim KS, Choi GH, Yun M. Prognostic value of metabolic tumor volume and total lesion glycolysis on preoperative 18F-FDG PET/CT in patients with very early and early hepatocellular carcinoma. Clin Nucl Med. 2017; 42:34–39. PMID: 27775949.

18. Schug ZT, Vande Voorde J, Gottlieb E. The metabolic fate of acetate in cancer. Nat Rev Cancer. 2016; 16:708–717. PMID: 27562461.

19. Ho CL, Yu SC, Yeung DW. 11C-acetate PET imaging in hepatocellular carcinoma and other liver masses. J Nucl Med. 2003; 44:213–221. PMID: 12571212.
20. Bjornson E, Mukhopadhyay B, Asplund A, Pristovsek N, Cinar R, Romeo S, et al. Stratification of hepatocellular carcinoma patients based on acetate utilization. Cell Rep. 2015; 13:2014–2026. PMID: 26655911.
21. Jeon J, Lee M, Whang S, Kim JW, Cho A, Yun M. Regulation of acetate utilization by monocarboxylate transporter 1 (MCT1) in hepatocellular carcinoma (HCC). Oncol Res. 2018; 26:71–81. PMID: 28390113.

22. Breuhahn K, Gores G, Schirmacher P. Strategies for hepatocellular carcinoma therapy and diagnostics: lessons learned from high throughput and profiling approaches. Hepatology. 2011; 53:2112–2121. PMID: 21433041.

23. Thorgeirsson SS. Genomic decoding of hepatocellular carcinoma. Gastroenterology. 2006; 131:1344–1346. PMID: 17030203.

24. Wang K, Lim HY, Shi S, Lee J, Deng S, Xie T, et al. Genomic landscape of copy number aberrations enables the identification of oncogenic drivers in hepatocellular carcinoma. Hepatology. 2013; 58:706–717. PMID: 23505090.

25. Kan Z, Zheng H, Liu X, Li S, Barber TD, Gong Z, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013; 23:1422–1433. PMID: 23788652.

26. Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato M, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014; 46:1267–1273. PMID: 25362482.

27. Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015; 47:505–511. PMID: 25822088.

28. Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012; 44:760–764. PMID: 22634756.

29. Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012; 44:694–698. PMID: 22561517.

30. Huang J, Deng Q, Wang Q, Li KY, Dai JH, Li N, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 2012; 44:1117–1121. PMID: 22922871.

31. Cleary SP, Jeck WR, Zhao X, Chen K, Selitsky SR, Savich GL, et al. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology. 2013; 58:1693–1702. PMID: 23728943.

32. Ahn SM, Jang SJ, Shim JH, Kim D, Hong SM, Sung CO, et al. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology. 2014; 60:1972–1982. PMID: 24798001.
33. Nault JC, Zucman-Rossi J. TERT promoter mutations in primary liver tumors. Clin Res Hepatol Gastroenterol. 2016; 40:9–14. PMID: 26336998.

34. Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009; 23:537–548. PMID: 19270154.

35. Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006; 126:107–120. PMID: 16839880.

36. Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009; 15:6479–6483. PMID: 19861459.

37. Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997; 94:6658–6663. PMID: 9192621.

38. Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010; 330:1340–1344. PMID: 21127244.

39. Méndez-Lucas A, Li X, Hu J, Che L, Song X, Jia J, et al. Glucose catabolism in liver tumors induced by c-MYC can be sustained by various PKM1/PKM2 ratios and pyruvate kinase activities. Cancer Res. 2017; 77:4355–4364. PMID: 28630053.

40. Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992; 12:5447–5454. PMID: 1448077.

41. Semenza GL, Roth PH, Fang HM, Wang GL. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol Chem. 1994; 269:23757–23763. PMID: 8089148.

42. Firth JD, Ebert BL, Pugh CW, Ratcliffe PJ. Oxygen-regulated control elements in the phosphoglycerate kinase 1 and lactate dehydrogenase A genes: similarities with the erythropoietin 3’ enhancer. Proc Natl Acad Sci U S A. 1994; 91:6496–6500. PMID: 8022811.

43. Rigiracciolo DC, Scarpelli A, Lappano R, Pisano A, Santolla MF, De Marco P, et al. Copper activates HIF-1α/GPER/VEGF signalling in cancer cells. Oncotarget. 2015; 6:34158–34177. PMID: 26415222.

44. Xia X, Lemieux ME, Li W, Carroll JS, Brown M, Liu XS, et al. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci U S A. 2009; 106:4260–4265. PMID: 19255431.

45. Hamaguchi T, Iizuka N, Tsunedomi R, Hamamoto Y, Miyamoto T, Iida M, et al. Glycolysis module activated by hypoxia-inducible factor 1alpha is related to the aggressive phenotype of hepatocellular carcinoma. Int J Oncol. 2008; 33:725–731. PMID: 18813785.

46. Daskalow K, Rohwer N, Raskopf E, Dupuy E, Kühl A, Loddenkemper C, et al. Role of hypoxia-inducible transcription factor 1alpha for progression and chemosensitivity of murine hepatocellular carcinoma. J Mol Med (Berl). 2010; 88:817–827. PMID: 20383692.
47. Lau CK, Yang ZF, Ho DW, Ng MN, Yeoh GC, Poon RT, et al. An Akt/hypoxia-inducible factor-1alpha/platelet-derived growth factor-BB autocrine loop mediates hypoxia-induced chemoresistance in liver cancer cells and tumorigenic hepatic progenitor cells. Clin Cancer Res. 2009; 15:3462–3471. PMID: 19447872.
48. Krishnamachary B, Zagzag D, Nagasawa H, Rainey K, Okuyama H, Baek JH, et al. Hypoxia-inducible factor-1-dependent repression of E-cadherin in von Hippel-Lindau tumor suppressor-null renal cell carcinoma mediated by TCF3, ZFHX1A, and ZFHX1B. Cancer Res. 2006; 66:2725–2731. PMID: 16510593.

49. Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008; 10:295–305. PMID: 18297062.
50. Zhang L, Huang G, Li X, Zhang Y, Jiang Y, Shen J, et al. Hypoxia induces epithelial-mesenchymal transition via activation of SNAI1 by hypoxia-inducible factor-1α in hepatocellular carcinoma. BMC Cancer. 2013; 13:108. PMID: 23496980.

51. Wang L, Wang WL, Zhang Y, Guo SP, Zhang J, Li QL. Epigenetic and genetic alterations of PTEN in hepatocellular carcinoma. Hepatol Res. 2007; 37:389–396. PMID: 17441812.

52. Peyrou M, Bourgoin L, Foti M. PTEN in liver diseases and cancer. World J Gastroenterol. 2010; 16:4627–4633. PMID: 20872961.
53. Rahman MA, Kyriazanos ID, Ono T, Yamanoi A, Kohno H, Tsuchiya M, et al. Impact of PTEN expression on the outcome of hepatitis C virus-positive cirrhotic hepatocellular carcinoma patients: possible relationship with COX II and inducible nitric oxide synthase. Int J Cancer. 2002; 100:152–157. PMID: 12115563.

54. Zundel W, Schindler C, Haas-Kogan D, Koong A, Kaper F, Chen E, et al. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev. 2000; 14:391–396. PMID: 10691731.

55. Tian T, Nan KJ, Wang SH, Liang X, Lu CX, Guo H, et al. PTEN regulates angiogenesis and VEGF expression through phosphatase-dependent and -independent mechanisms in HepG2 cells. Carcinogenesis. 2010; 31:1211–1219. PMID: 20430845.

56. Chen C, Pore N, Behrooz A, Ismail-Beigi F, Maity A. Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. J Biol Chem. 2001; 276:9519–9525. PMID: 11120745.
57. Sakamoto T, Niiya D, Seiki M. Targeting the Warburg effect that arises in tumor cells expressing membrane type-1 matrix metalloproteinase. J Biol Chem. 2011; 286:14691–14704. PMID: 21372132.

58. Wang H, Zhang C, Xu L, Zang K, Ning Z, Jiang F, et al. Bufalin suppresses hepatocellular carcinoma invasion and metastasis by targeting HIF-1α via the PI3K/AKT/mTOR pathway. Oncotarget. 2016; 7:20193–20208. PMID: 26958938.

59. Wang Y, Ren J, Gao Y, Ma JZ, Toh HC, Chow P, et al. MicroRNA-224 targets SMAD family member 4 to promote cell proliferation and negatively influence patient survival. PLoS One. 2013; 8:e68744. PMID: 23922662.

60. Wang Y, Lee CG. MicroRNA and cancer--focus on apoptosis. J Cell Mol Med. 2009; 13:12–23. PMID: 19175697.
61. Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008; 27:5651–5661. PMID: 18521080.

62. Lee J, Kemper JK. Controlling SIRT1 expression by microRNAs in health and metabolic disease. Aging (Albany NY). 2010; 2:527–534. PMID: 20689156.

63. Lee J, Padhye A, Sharma A, Song G, Miao J, Mo YY, et al. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J Biol Chem. 2010; 285:12604–12611. PMID: 20185821.

64. Qin Y, Lu Y, Wang R, Li W, Qu X. SL1122-37, a novel derivative of sorafenib, has greater effects than sorafenib on the inhibition of human hepatocellular carcinoma (HCC) growth and prevention of angiogenesis. Biosci Trends. 2013; 7:237–244. PMID: 24241174.

65. Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008; 359:378–390. PMID: 18650514.

66. Aft RL, Zhang FW, Gius D. Evaluation of 2-deoxy-D-glucose as a chemotherapeutic agent: mechanism of cell death. Br J Cancer. 2002; 87:805–812. PMID: 12232767.

67. Zhang Y, Huang F, Wang J, Luo H, Wang Z. 2-DG-regulated RIP and c-FLIP effect on liver cancer cell apoptosis induced by TRAIL. Med Sci Monit. 2015; 21:3442–3448. PMID: 26552967.

68. Tesori V, Piscaglia AC, Samengo D, Barba M, Bernardini C, Scatena R, et al. The multikinase inhibitor Sorafenib enhances glycolysis and synergizes with glycolysis blockade for cancer cell killing. Sci Rep. 2015; 5:9149. PMID: 25779766.

69. Geschwind JF, Georgiades CS, Ko YH, Pedersen PL. Recently elucidated energy catabolism pathways provide opportunities for novel treatments in hepatocellular carcinoma. Expert Rev Anticancer Ther. 2004; 4:449–457. PMID: 15161443.

70. Ko YH, Verhoeven HA, Lee MJ, Corbin DJ, Vogl TJ, Pedersen PL. A translational study “case report” on the small molecule “energy blocker” 3-bromopyruvate (3BP) as a potent anticancer agent: from bench side to bedside. J Bioenerg Biomembr. 2012; 44:163–170. PMID: 22328020.

71. Schimmer AD, Thomas MP, Hurren R, Gronda M, Pellecchia M, Pond GR, et al. Identification of small molecules that sensitize resistant tumor cells to tumor necrosis factor-family death receptors. Cancer Res. 2006; 66:2367–2375. PMID: 16489043.

72. Wood TE, Dalili S, Simpson CD, Hurren R, Mao X, Saiz FS, et al. A novel inhibitor of glucose uptake sensitizes cells to FAS-induced cell death. Mol Cancer Ther. 2008; 7:3546–3555. PMID: 19001437.

73. Liu Y, Cao Y, Zhang W, Bergmeier S, Qian Y, Akbar H, et al. A small-molecule inhibitor of glucose transporter 1 downregulates glycolysis, induces cell-cycle arrest, and inhibits cancer cell growth in vitro and in vivo. Mol Cancer Ther. 2012; 11:1672–1682. PMID: 22689530.
Fig. 1
Hepatocellular carcinoma positive for F-18 fluorodeoxyglucose (A), but negative for C-11 acetate (B).
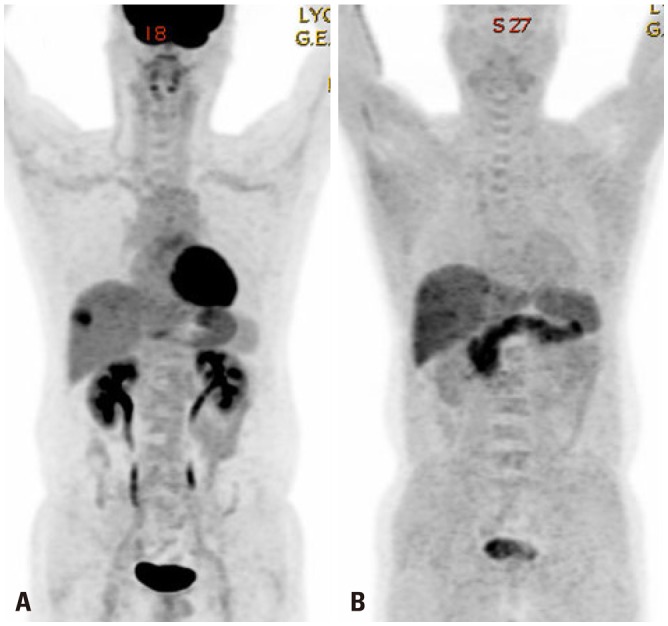
Fig. 2
Hepatocellular carcinoma negative for F-18 fluorodeoxyglucose (A), but positive C-11 acetate (B).
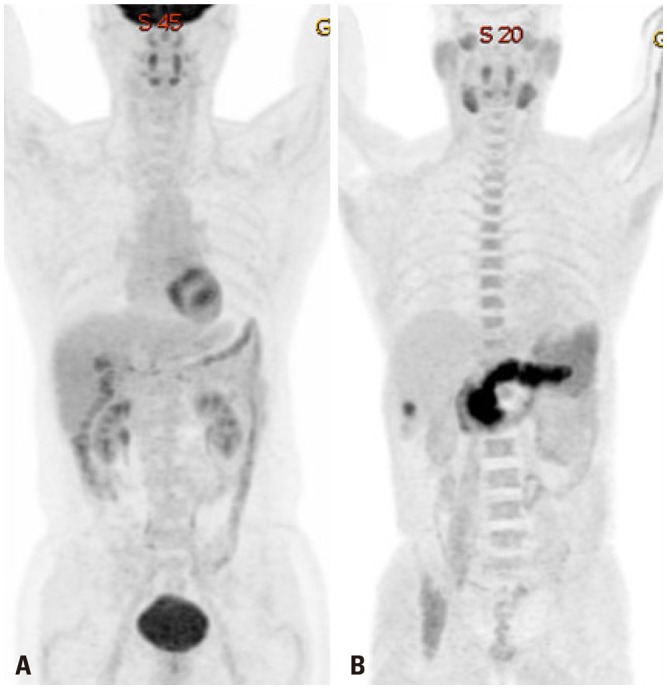
Fig. 3
Differences in the expression of glucose transport 1 (A and C) and monocarboxylate transporter 1 (B and D) in hepatocellular carcinoma (HCC) samples, based on 18F-fluorodeoxyglucose and 11C-acetate uptake. Human HCC samples were used. Immunohistochemistry (IHC) was performed as described previously.16 After antigen retrieval, IHC was performed using indicated antibodies. Scale bars: 40 µm.
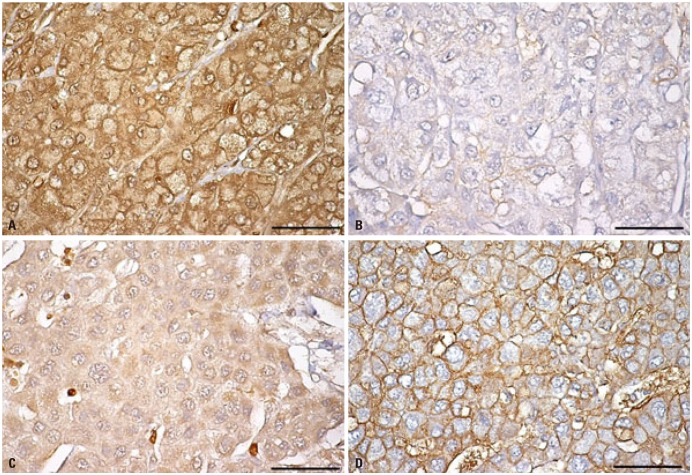
Table 1
List of the Most Relevant Mutations in Hepatocellular Carcinoma




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download