Abstract
In order to investigate the occurrence of norovirus in rivers and beaches, a total of 81 samples were tested at seven sites of Oncheon stream, Suyeong river and Gwanganri beach in Busan from January to November, 2017. To improve the detection of norovirus from sea water, we applied the inorganic cation-coated filter method which showed 48.8% ± 12.2% (n=3) and 27.4% ± 6.0% (n=3) recovery yields from river water and sea water inoculated with Norovirus, respectively. Norovirus was detected in a total of four samples (4.9%), which all were GII genotype. Norovirus GII was detected in three samples at two waste water treatment plants (WWTP) outlet and one sample at about 500 meter downstream from WWTP in both the winter and spring seasons. We also monitored fecal indicator organisms, Escherichia coli (E. coli), Enterococcus and coliphages [somatic coliphages (SC), male-specific coliphages (MSC)] to analyze the potential transmission of enteritis causative agent in dry and wet days. Bacterial influences were found at the site of the WWTP effluents in the dry days and spread further to the costal beach in the wet days. But no viral influences were found in the river downstream in both dry and wet days.
REFERENCES
1). Belliot G, Noel JS, Li JF, Seto Y, Humphrey CD, Ando T, et al. Characterization of capsid genes, expressed in the baculovirus system of three new genetically distinct strains of Norwalk-like viruses. J Clin Microbiol. 2001; 39:4288–95.

2). Green J, Wright PA, Gallimore CI, Mitchell O, Morgan Capner P, Brown DW. The role of environmental contamination with small round structured viruses in a hospital outbreak investigated by reverse-transcriptase polymerase chain reaction assay. J Hosp Infect. 1998; 39:39–45.

3). Zheng DP, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS. Norovirus classification and proposed strain nomenclature. Virology. 2006; 346:312–23.

4). Jung JH, Yoo CH, Koo ES, Kim HM, Na Y, Jheong WH, et al. Occurence of norovirus and other enteric viruses in untreated groundwaters of Korea. J Water Health. 2011; 9:544–55.
5). La Rosa G, Fontana S, Di Grazia A, Iaconelli M, Pourshaban M, Muscillo M. Molecular identification and genetic analysis of Norovirus genogroups Ⅰand Ⅱin water environments: comparative analysis of different reverse transcription-PCR assays. Appl Environ Microbiol. 2007; 73:4152–61.
6). Kroneman A, Verhoef L, Harris J, Vennerma H, Duizer E, van Duynhoven Y, et al. Analysis of integrated virological and epidemiological reporter of norovirus outbreaks collected within the foodborne viruses in Europe network from 1 July 2001 to 30 June 2006. J Clin Microbiolol. 2008; 46:2959–65.
7). Yang N, Qi H, Wong MM, Wu RS, Kong RY. Prevalence and diversity of norovirus genogroups Ⅰand Ⅱin Hong Kong marine waters and detection by real-time PCR. Mar Pollt Bull. 2012; 64:164–8.
8). Katayama H, Haramoto E, Oguma K, Yamashita H, Tajima A, Nakajima H, et al. One-year monthly quantitative survey of noroviruses, enteroviruses, and adenoviruses in wastewater collected from six plants in Japan. Water Res. 2008; 42:1441–8.

9). Rodriguez-Diaz J, Querales L, Caraballo L, Vizzi E, Liprandi F, Takiff H, et al. Detection and characterization of waterborne gastroenteritis viruses in urban sewage and sewage-polluted river waters in Caracas, Venezuela. Appl Environ Microbiol. 2009; 75:387–94.

10). Aw TG, Gin KY. Environmental surveilance and molecular characterization of human enteric viruses in tropical urban wastewaters. J Appl Microbiol. 2010; 109:716–30.
11). Ueki Y, Sano D, Watanabe T, Akiyama K, Omura T. Norovirus pathway in water environment estimated by genetic analysis of strains from patients of gastroenteritis, sewage, treated wastewater, river water and oyster. Water Res. 2005; 39:4271–80.
12). Lee C, Kim SJ. The genetic diversity of human noroviruses detected in river water in Korea. Water Res. 2008; 42:4477–84.

13). Gentry J, Vinjé J, Guadagnoli D, Lipp EK. Norovirus distribution within an estuarine environment. Appl Environ Microbiol. 2009; 75:5474–80.

14). Kitajima M, Haramoto E, Phanuwan C, Katayama H, Ohgaki S. Detection of genogruoup Ⅳin wastewater and river water in Japan. Lett Appl Microbiol. 2009; 49:655–8.
15). Mans J, Netshikweta R, Magwalivha M, Van Zyl WB, Taylor MB. Diverse norovirus genotypes identified in sewage-polluted river water in South Africa. Epidemiol Infect. 2013; 141:303–13.

16). Katayama H, Shimasaki A, Ohgaki S. Development of a virus concentration method and its application to detection of enterovirus and norwalk virus from coastal seawater. Appl Environ Microbiol. 2002; 68:1033–9.

17). Tong HI, Connell C, Boehm AB, Lu Y. Effective detection of human noroviruses in Hawaiian waters using enhanced RT-PCR methods. Water Res. 2011; 45:5837–48.

18). Wyn-Jones AP, Carducci A, Cook N, D'Agostino M, Divizia M, Fleischer J, et al. Surveillance of adenovirus and noroviruses in European recreational waters. Water Res. 2011; 45:1025–38.
19). Gibbons CD, Rodriguez RA, Tallon L, Sobsey MD. Evaluation of positively charged alumina nanofibre cartridge filters for the primary concetration of norovirus, adenovirues and male-specific coliphages from seawater. J Appl Mircobiol. 2010; 109:635–41.
20). Abbaszadegan M, Huber MS, Gerba CP, Pepper IL. Detection of enteroviruses in groundwater with the polymerase chain reaction. Appl Environ Microbiol. 1993; 59:1318–24.

21). Jee BY, Kim KI, Lee SJ, Kim KH, Jin JW, Jeong HD. Detection of fish pathogenic viruses in seawater using negatively charged membranes. Korean J Fish Aquat Sci. 2013; 46:46–52.

22). Lee HJ, Oh EG, Yu HS, Shin SB, Son MJ, Jung JY, et al. Recovery of Norovirus surrogate in seawater using an electropositive and electronegative filter. Korean J Fish Aquat Sci. 2009; 42:238–42.

23). Koo HS, Ku PT, Lee MO, Baik HS. Prevalence of Noroviruses Detected from Outbreaks of Acute Gastroenteritis in Busan, Korea. J Life Sci. 2016; 26:911–20.

24). Jheong WH, Kim JM, Jang SJ, Park JY, Oh JH, Choi HJ, et al. A study on the bacteriophages as indicators of viruses in water environment. National Instittute of Environmental Research Report 2008-52-1002.
25). Grabow WOK. Microbiology of drinking water treatment; reclaimed wastewater. McFeters GA, editor. Drinking Water Microbiology-Progress and Recent Developments. New York: Springer Verlag;1990. p. 185–203.

26). Havelaar AH, Van olphen M, Drost YC. F-specific RNA bacteriophages are adequate model organisms for enteric viruses in fresh water. Appl Environ Microbiol. 1993; 59:2956–62.

27). AWPRC study group on health related water microbiology. Bacteriophages as model viruses in water quality control. Water Res. 1991; 25:529–45.
Figure 1.
Location of the sample collection sites. Location of the sites in which water samples were collected was indicated as S1~S7. S, site.
Figure 2.
Seasonal variation in the distribution of microorganism indicators at all study sites. E. coli, Escherichia coli; SC, Somatic coliphage; MSC, Male-specific coliphage. Error bars indicate standard deviation and E. coli (a) and SC (b) are showed significant difference between summer and winter according to t-test (p<0.05).
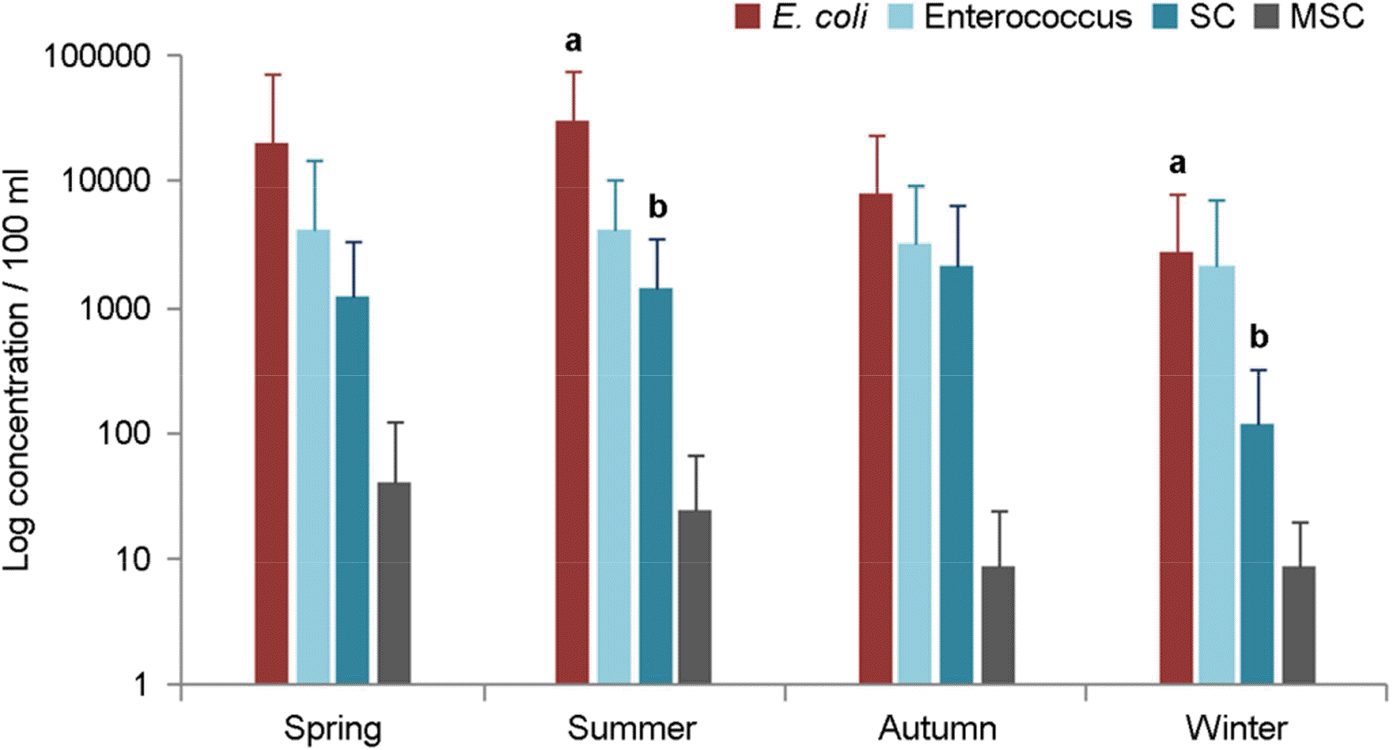
Figure 3.
Comparison of fecal bacteria and coliphages in dry days and wet days by sample location. It showed significant difference between Dry days and Wet days according to t-test (p<0.05).
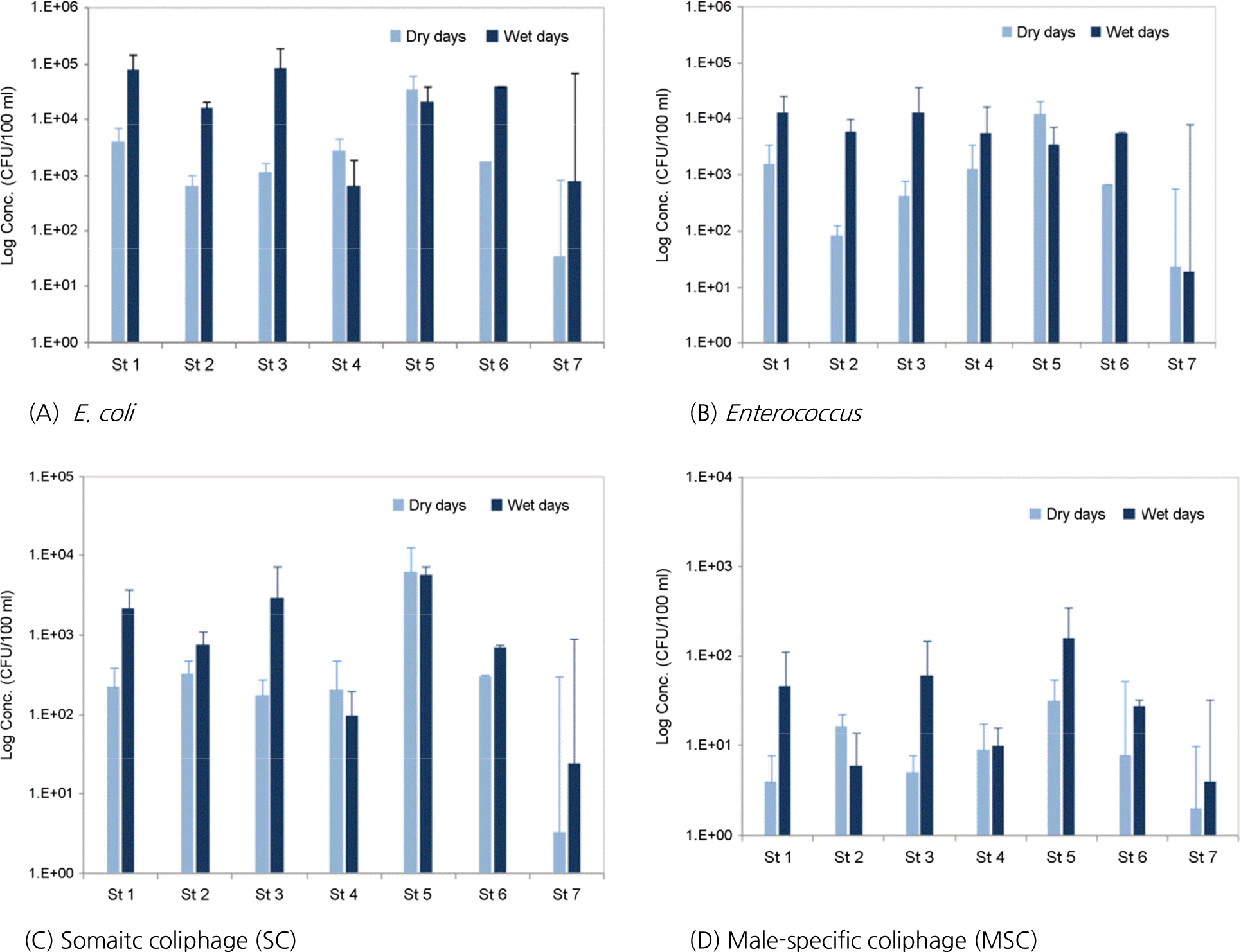
Figure 4.
Variability of microorganism indicator in dry days and wet days at study area. The color shows the changes of concentration level of the indicator microorganism. Concentration units are shown above each color bars.
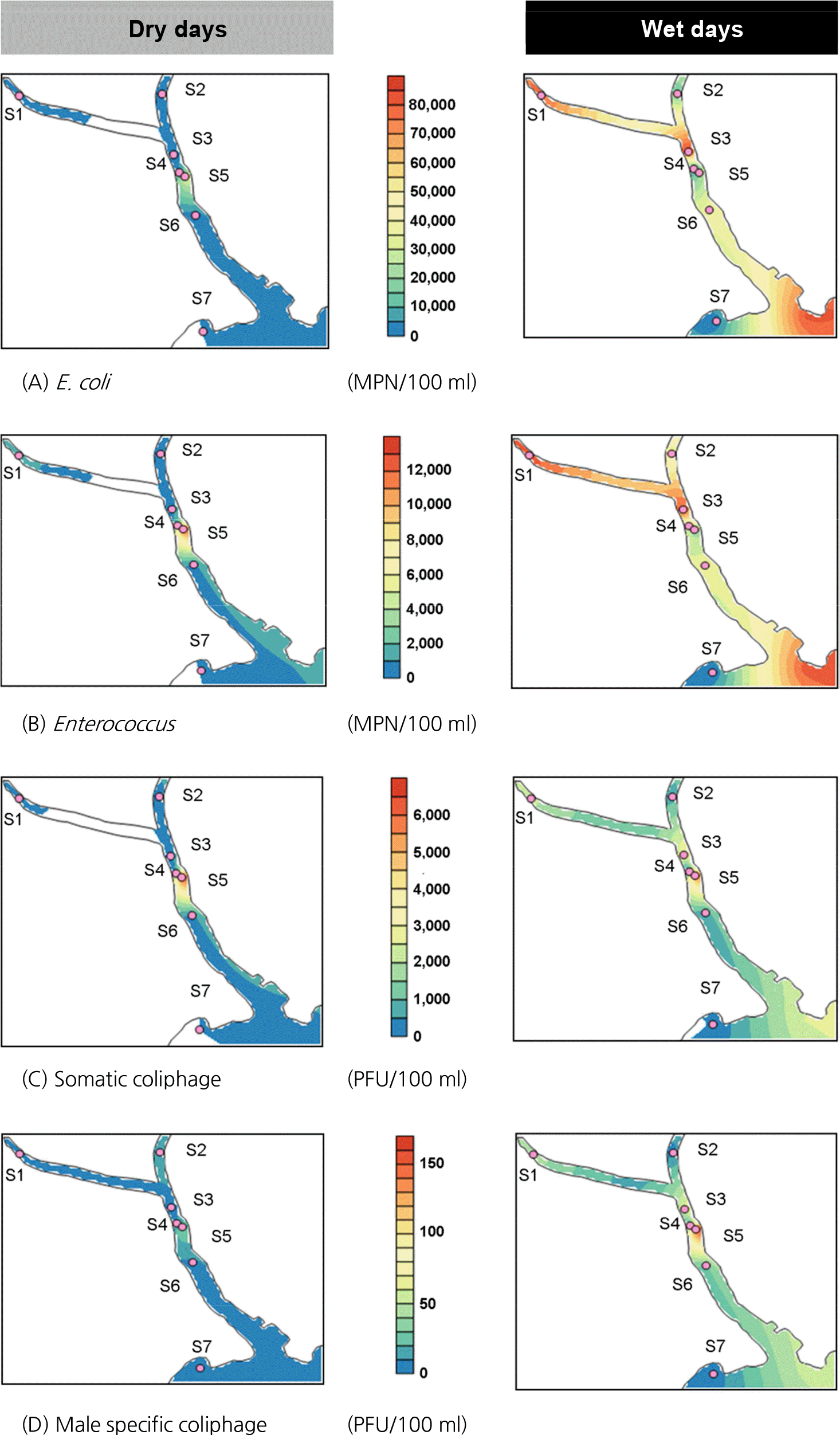
Table 1.
Characteristics of host cells of coliphages
Table 2.
The sequences of oligonucleotides used for the detection of norovirus
Table 3.
Recovery of Norovirus from natural seawater and stream water spiked with Norovirus GII
| Sample | Initial concentration (total 300 mL water) viral copies/ml | Recovered concentration (total 1 mL concentrate) viral copies/ml | Recovery ratea (%±SD) |
|---|---|---|---|
| Seawaterb | 6.9 × 104 | 1.9 ± 0.4 × 104 | 27.4 ± 6.0 |
| Stream waterb | 6.9 × 104 | 3.4 ± 0.8 × 104 | 48.8 ± 11.2 |
Table 4.
Parameter characteristics of norovirus (NoV) GII positive samples
Table 5.
Seasonal characteristics in physiochemical factors and waterborne microbial indicators
| Season | N | Temp. (℃) | pH | DO (mg/l) | Salinity (PSU) | Turbidity (NTU) | E. coli (MPN/ 100 ml) | Enterococcus (MPN/ 100 ml) | Coliphage (PFU/100 ml) | |
|---|---|---|---|---|---|---|---|---|---|---|
| SC | MSC | |||||||||
| Spring | 31 | 16.5 | 7.2 | 8.0 | 11.63 | 3.17 | 2.0×103 | 4.2×103 | 1.2×103 | 4.0×101 |
| (Dry days)a | (19) | 16.9 | 7.2 | 7.8 | 11.68 | 2.40 | 5.8×103 | 1.0×103 | 4.8×102 | 1.0×101 |
| (Wet days)b | (12) | 15.8 | 7.3 | 8.4 | 11.56 | 4.40 | 4.2×104 | 9.3×103 | 2.3×103 | 7.5×101 |
| Summer (Wet days)c | 14 | 23.7 | 7.2 | 6.6 | 13.88 | 2.75 | 3.0×104 | 4.2×103 | 1.4×103 | 2.5×101 |
| Autumn (Dry days) | 21 | 18.1 | 7.3 | 7.6 | 14.12 | 2.30 | 8.2×103 | 3.3×103 | 2.1×103 | 9.0×100 |
| Winter (Dry days) | 15 | 9.5 | 7.4 | 10.9 | 14.07 | 2.07 | 2.8×103 | 2.1×103 | 1.2×102 | 9.0×100 |
| Total | 81 | 16.9 | 7.3 | 8.2 | 13.10 | 2.7 | 1.6×104 | 3.6×103 | 1.3×103 | 2.2×101 |
Temp, Temperature; DO, Dissolved oxygen; PSU, Practical salinity unit; NTU, Nephelometric turbidity unit; E. coli, Escherichia coli; SC, Somatic coliphage; MSC, Male-specifc (F+) coliphage; MPN, most probable number; PFU, plaque forming unit; N, number of samples; Spring months: March, April, and May; Summer months: June, July, and August; Autumn months: September, October, and November; Winter months: December, January, and February.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download