Abstract
Background
The accurate identification of cytogenetic abnormalities in multiple myeloma (MM) has become more important over recent years for the development of new diagnostic and prognostic markers. In this study, we retrospectively analyzed the cytogenetic aberrations in MM cases as an initial assessment in a single institute.
Methods
We reviewed the cytogenetic results from 222 patients who were newly diagnosed with MM between January 2000 and December 2015. Chromosomal analysis was performed on cultured bone marrow samples by standard G-banding technique. At least 20 metaphase cells were analyzed for karyotyping.
Results
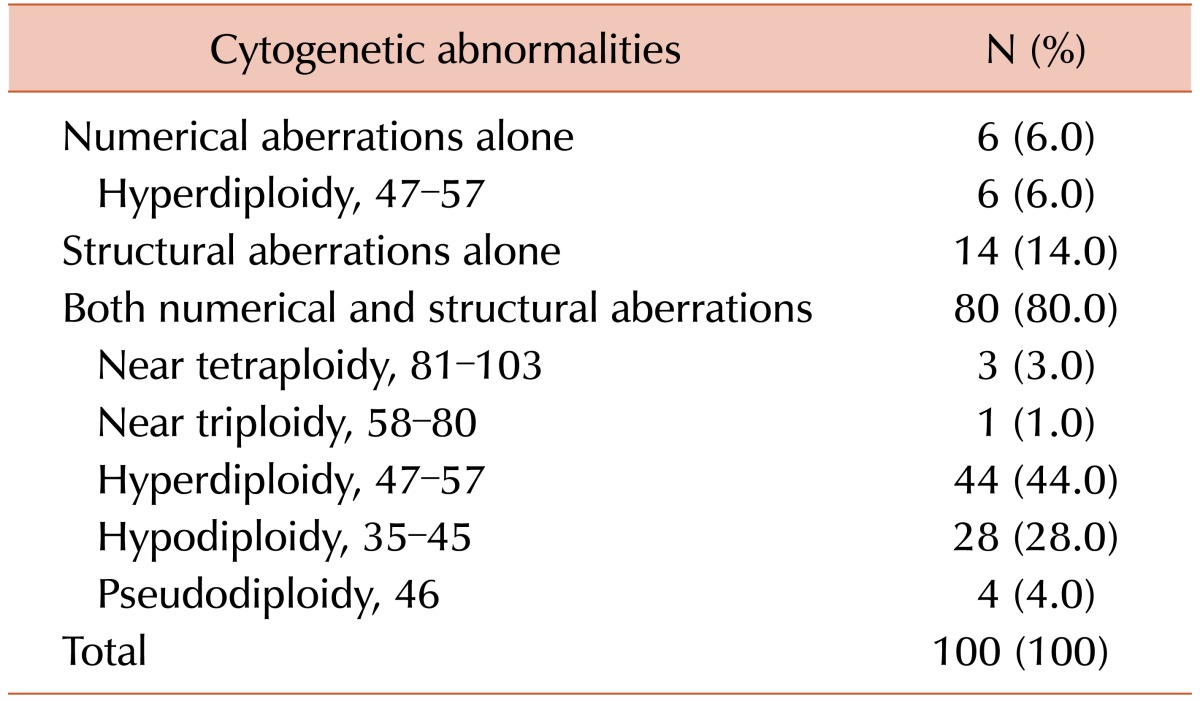
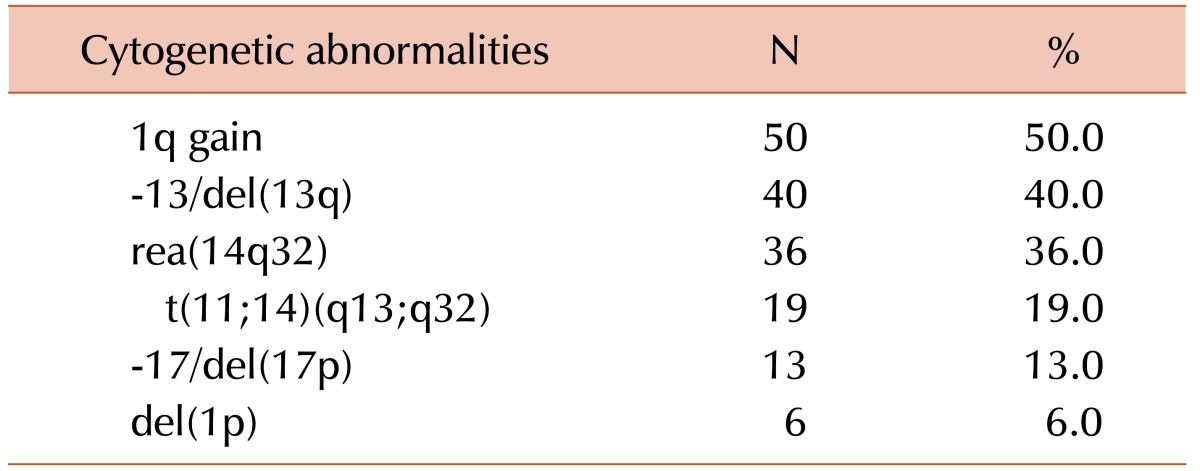
Clonal chromosome abnormalities were detected in 45.0% (100/222) of the patients. Among these results, 80 cases (80.0%) had both numerical and structural chromosome abnormalities. Overall hyperdiploidy with structural cytogenetic aberrations was the most common finding (44.0%), followed by hypodiploidy with structural aberrations (28.0%). Amplification of the long arm of chromosome 1 and -13/del(13q) were the most frequent recurrent abnormalities, and were detected in 50 patients (50.0%) and 40 patients (40.0%) with clonal abnormalities, respectively. The most common abnormality involving 14q32 was t(11;14)(q13;q32), which was observed in 19 cases.
Multiple myeloma (MM) is a plasma cell malignancy characterized by the accumulation of clonal plasma cells in the bone marrow. It is a heterogeneous disorder that progresses from a premalignant stage called monoclonal gammopathy of undetermined significance (MGUS) to symptomatic myeloma with bone destruction, suppression of bone marrow function, and renal insufficiency [12]. The International Myeloma Working Group (IMWG) has established the diagnostic criteria for plasma cell disorders [34].
Conventional cytogenetic analysis should be included in the initial diagnostic work-up for patients suspected of having MM. Cytogenetic alterations in MM represent important risk factors in terms of prognosis. Abnormal karyotypes are found in 30–50% of cases, more often in advanced-stage patients than in newly diagnosed patients [56]. While cytogenetic analyses can provide useful prognostic information, the low spontaneous proliferative activity of the tumor cells, especially early stage of the disease, is considered to be a significant limiting factor. Interpretation can be difficult, even within abnormal metaphase cells, because some aberrations are cryptic, and chromosomal morphologies can be of poor quality [567]. These limitations have been partly overcome by the use of molecular cytogenetic techniques such as fluorescence in situ hybridization (FISH), and comparative genomic hybridization (CGH). However, the FISH approach involves only targeting selected genes, and CGH cannot detect some balanced rearrangements. In this study, we retrospectively analyzed the cytogenetic aberrations in 222 patients with newly diagnosed MM, as an initial assessment at a single institute.
This study enrolled 222 patients newly diagnosed with MM between January 2000 and December 2015. The criteria for the diagnoses were based on the IMWG definition [34]. The morphology of patient bone marrow specimens confirmed myeloma in all cases. Informed consents were obtained from all patients prior to the study.
Fresh bone marrow aspirate samples were cultured as 48- and 72-hour unstimulated cultures following standard cytogenetic methods. To increase the mitotic index of the cultures, stimulation with 2 µg/mL phorbol 12-myristate 13-acetate and 200 µL/mL phytohaemagglutinin was also used. Chromosomal analysis was performed on cultured bone marrow samples using the standard G-banding technique. At least 20 metaphase cells were used for karyotyping, and results were reported according to the International System for Human Cytogenetic Nomenclature (2013) [8].
Identification of clonal abnormalities involving chromosome gain or structural rearrangement required that at least two cells had the same aberration. Identification of clonal abnormalities involving loss of a chromosome required the presence of at least three cells with the same chromosome loss. In terms of the modal number, hyperdiploidy was used to describe cells with 47–57 chromosomes, and hypodiploidy was used to describe cells with 35–45 chromosomes. Near triploidy was defined as 58–80 chromosomes, near tetraploidy as 81–103 chromosomes, and pseudodiploidy was defined as 46 chromosomes with numerical and/or structural aberrations [8]. A complex karyotype was defined as having three or more chromosomal abnormalities, including at least one structural aberration. This project was approved by Dong-A University Hospital Institutional Review Board. Descriptive statistics were used to summarize the cytogenetic aberrations.
Out of 222 patients, 119 (54.1%) were male and the male to female gender ratio was 1.16. The median age at diagnosis was 63.3 years, and ages ranged from 42 to 91. The percentage of marrow plasma cells varied between 10.5 and 91.0. There were 114 cases of immunoglobulin isotype IgG (51.5%), 34 cases of IgA (15.2%), and eight cases of IgM (3.5%). Light chain isotypes were observed in 66 patients (29.8%), of which 38 (57.8%) were kappa (κ) and 28 (42.2%) were lambda (λ). Clinical staging using the International Staging System (ISS) [9] was possible in 213 patients. The number of patients in stages I, II, and III were 10, 45, and 158, respectively.
Chromosome abnormalities were detected in 45.0% (100/222) of the patients (Table 1). Among these results, 80 cases (80.0%) had both numerical and structural chromosome abnormalities. Fourteen patients (14.0%) had structural abnormalities alone and 6 (6.0%) had numerical abnormalities alone, of which all included hyperdiploidy. Overall hyperdiploidy with structural cytogenetic aberrations was the most common finding (44.0%), followed by hypodiploidy with structural cytogenetic aberrations (28.0%). Almost all of the latter two groups showed complex karyotypes (100% and 92.9%, respectively).
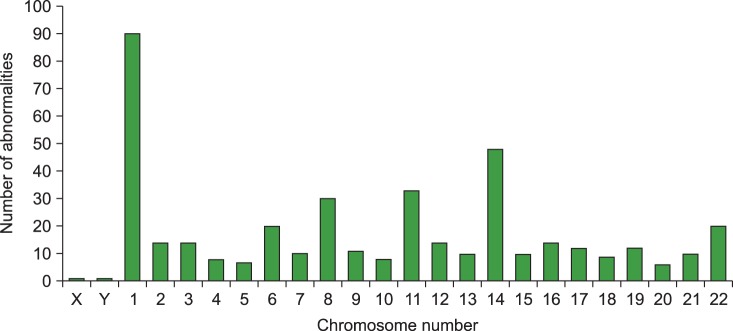
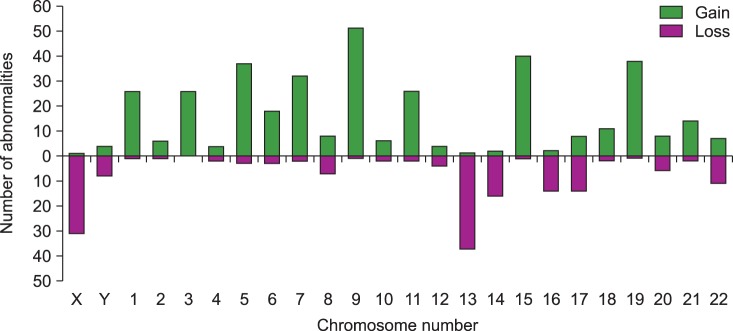
The most common abnormalities were structural aberrations and gains involving chromosome 1 (117/963 total abnormalities; 12.1%), which were frequently identified as unbalanced translocation resulting in partial trisomy 1q (Fig. 1, 2). The next most common abnormalities were structural aberrations with loss of chromosome 14 (6.9%), chromosome 9 (6.5%), and chromosome 11 (6.3%). Chromosome gain was most commonly observed with chromosome 9 (51/380 gains; 13.4%), followed by chromosomes 15, 19, 5, 7, and 3 (Fig. 2). Loss of chromosome 13 (37/171 losses; 21.6%) was most common among monosomies, followed by loss of the X chromosome, and then by loss of chromosomes 14, 16, and 17.
Amplification of the long arm of chromosome 1 was the most frequent recurrent abnormality, which was detected in 50 patients (50.0%) with clonal abnormalities (Table 2). As the second most common abnormality, -13/del(13q) was observed in 40 cases (40.0%). Twenty-five of 50 cases of 1q gain (50.0%) had concurrent -13/del(13q) aberrations. Chromosome 14q32 abnormalities were found in 36 cases (36.0%). The most common abnormality involving 14q32 was t(11;14)(q13;q32), which was observed in 19 cases. We also found rearrangements of 14q32 other than t(11;14) in 17 patients, of which add(14q32) abnormalities were seen in 11, t(8;14)(q24.1;q32) in four, and t(3;14)(p21;q32) and t(10;14)(q22;q32) in one patient each. Eight of 17 cases with t(11;14) (42.1%) also had concurrent 1q gain. Monosomy 17 or 17p deletions and chromosome 1p deletions were found in 13 and six patients, respectively.
Traditional approaches for the genetic characterization of multiple myelomas include cytogenetic analysis, FISH, and molecular genetic studies. These technologies are complementary to each other and provide important diagnostic and prognostic information. As new insights into the complexities and heterogeneities of myelomas at the molecular level are obtained, the need emerges to attain a more complete molecular genetic profile. With the advent of next-generation sequencing (NGS), a more detailed biological characterization of myelomas can be achieved at the molecular level [12101112].
Cytogenetic information can be limited, as the malignant plasma cell precursors might have low mitotic frequencies. Some aberrations, such as t(4;14)(p16;q32), are cryptic and cannot be detected with conventional banding techniques. Of 222 eligible MM patients in the present study, 100 (45.0%) had clonal chromosome abnormalities. Within the published literature, abnormal karyotypes are reported to be found in 30–50% of cases when traditional cytogenetic analysis is applied, whereas they vary widely between 15.0% and 75.6% when real analytical data is examined [56101113141516]. These different detection rates can arise from multiple factors such as tumor heterogeneity, the stages of the patients studied, the cell-culture techniques used to obtain metaphase cells, and variations in the interpretation of the data. Our observed abnormality rate was high, although lower than the previously obtained rate (57.1%) from the same institution [17].
Overall, hyperdiploidy with structural cytogenetic aberration was the most common finding (44.0%). Amplification of the long arm of chromosome 1 was the most frequent abnormality, and was detected in 50 patients (50.0%) with clonal chromosome abnormalities, consistent with previous reports [25610]. A second common abnormality was monosomy 13 or del(13q), which was observed in 40 cases (40.0%). Fifty percent of 1q gain cases had -13/del(13q) aberrations simultaneously. Some studies have previously reported monosomy 13 or del(13q) as the most common abnormality [11151618], and the frequency of this was even higher when FISH analysis was used. However, since there have been FISH studies that found more 1q abnormalities than -13/del(13q) abnormalities [1920], we need to pay attention to the composition of the panel of FISH probes employed for the study. In addition, because chromosome 1q abnormalities are often complex and can include various forms of duplications, whole arm translocations, isochromosomes, unbalanced derivative chromosomes, and jumping translocations, careful examinations of metaphase cells are also necessary. Secondary aberrations such as 1q gain or del(1p), -13/del(13q), and -17/del(17p) are all known to be important in the clonal expansion of MM [25621]. The minimal common regions of the aberrations are 1q21-q23.3, 13q14.3, and 17p13. The implicated genes are IL6R, CKS1B, and ANP32E in 1q, RB1 in 13q, and TP53 in 17p.
Primary translocations are believed to occur early in the pathogenesis of MM. The most frequent translocation is t(11;14)(q13;q32), which is found in about 15% of MM cases. The second most frequent translocation is t(4;14)(p16;q32), but it is cryptic and only detectable by FISH or other sensitive molecular methods [256711151618192021]. These translocation abnormalities are reported to be associated with up-regulation of one of the cyclin D genes. In our study, t(11;14)(q13;q32) was the most common translocation, occurring in 19 cases (19.0%). Using conventional G-banding alone, we could not find any t(4;14)(p16;q32) abnormalities. Instances of unidentifiable extra genetic material on the long arm of chromosome 14 in the form of add(14q32) abnormalities were observed in 11 cases. Four cases of t(8;14) (q24.1;q32) were found, and they were part of a complex karyotype, having t(11;14)(q13;q32) simultaneously in two patients. Translocations involving MYC are considered to be later events in MM tumor progression.
Gains of chromosomes most commonly involved chromosome 9 (51 of 380 gains; 13.4%), followed by chromosomes 15, 19, 5, 7, and 3. Among the chromosome loss, monosomy 13 was most frequent (37 of 171 losses; 21.6%), followed by X chromosome loss, and loss of chromosomes 14, 16, and 17. The frequencies of chromosome gains and losses in the present study were similar to those of other reports [256111216]. Currently, trisomies and IGH abnormalities are regarded as primary abnormalities, developing from the early stage of MM. Research to understand the role and mechanisms of trisomy in myeloma patients with major risk abnormalities such as t(4;14)(p16;q32) or del(17p) is still ongoing [122223].
Conventional cytogenetic studies in hematologic malignancies provide the advantage of whole genome analysis in one experiment. Using FISH increases the number of cells analyzed, and allows detection of submicroscopic abnormalities; however, FISH only targets selected genes and may be expensive when large panels of probes are used. In the present study, although we did not introduce FISH data to compare with conventional karyotyping, we could provide comprehensive cytogenetic analyses. These findings demonstrate that with a higher rate of cytogenetic abnormality detection, myeloma cells exhibit complex and heterogeneous aberrations regardless of ploidy.
In summary, our cytogenetic analyses showed a higher abnormality rate than other previous studies performed in Korea and enabled comprehensive detection of genetic changes in MM cases. However, further prospective studies are needed to identify high-risk genetic profiles and allow new risk-adapted diagnoses and treatments for MM patients.
References
1. Rajkumar SV. Multiple myeloma: 2014 Update on diagnosis, risk-stratification, and management. Am J Hematol. 2014; 89:999–1009. PMID: 25223428.

2. Talley PJ, Chantry AD, Buckle CH. Genetics in myeloma: genetic technologies and their application to screening approaches in myeloma. Br Med Bull. 2015; 113:15–30. PMID: 25662536.

3. Munshi NC, Anderson KC, Bergsagel PL, et al. Consensus recommendations for risk stratification in multiple myeloma: report of the International Myeloma Workshop Consensus Panel 2. Blood. 2011; 117:4696–4700. PMID: 21292777.

4. Rajkumar SV, Dimopoulos MA, Palumbo A, et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014; 15:e538–e548. PMID: 25439696.
6. Sawyer JR. The prognostic significance of cytogenetics and molecular profiling in multiple myeloma. Cancer Genet. 2011; 204:3–12. PMID: 21356186.

7. Jimenez-Zepeda VH, Braggio E, Fonseca R. Dissecting karyotypic patterns in non-hyperdiploid multiple myeloma: an overview on the karyotypic evolution. Clin Lymphoma Myeloma Leuk. 2013; 13:552–558. PMID: 23856591.

8. Shaffer LG, McGowan-Jordan J, Schmid M. An international system for human cytogenetic nomenclature. Basel, Switzerland: S Karger;2013.
9. Greipp PR, San Miguel J, Durie BG, et al. International staging system for multiple myeloma. J Clin Oncol. 2005; 23:3412–3420. PMID: 15809451.

10. Lai YY, Huang XJ, Cai Z, et al. Prognostic power of abnormal cytogenetics for multiple myeloma: a multicenter study in China. Chin Med J (Engl). 2012; 125:2663–2670. PMID: 22931972.
11. Lim AS, Lim TH, See KH, et al. Cytogenetic and molecular aberrations of multiple myeloma patients: a single-center study in Singapore. Chin Med J (Engl). 2013; 126:1872–1877. PMID: 23673102.
12. Kumar S, Fonseca R, Ketterling RP, et al. Trisomies in multiple myeloma: impact on survival in patients with high-risk cytogenetics. Blood. 2012; 119:2100–2105. PMID: 22234687.

13. Berry NK, Bain NL, Enjeti AK, Rowlings P. Genomic profiling of plasma cell disorders in a clinical setting: integration of microarray and FISH, after CD138 selection of bone marrow. J Clin Pathol. 2014; 67:66–69. PMID: 23969274.

14. Waheed S, Shaughnessy JD, van Rhee F, et al. International staging system and metaphase cytogenetic abnormalities in the era of gene expression profiling data in multiple myeloma treated with total therapy 2 and 3 protocols. Cancer. 2011; 117:1001–1009. PMID: 20945320.

15. Jekarl DW, Min CK, Kwon A, et al. Impact of genetic abnormalities on the prognoses and clinical parameters of patients with multiple myeloma. Ann Lab Med. 2013; 33:248–254. PMID: 23826560.

16. Lim JH, Seo EJ, Park CJ, et al. Cytogenetic classification in Korean multiple myeloma patients: prognostic significance of hyperdiploidy with 47-50 chromosomes and the number of structural abnormalities. Eur J Haematol. 2014; 92:313–320. PMID: 24372944.

17. Kim SH, Kim JH, Lee DM, et al. Comparison between conventional cytogenetics and interphase fluorescence in situ hybridization (FISH) for patients with multiple myeloma. Korean J Hematol. 2009; 44:14–21.

18. Jimenez-Zepeda VH, Neme-Yunes Y, Braggio E. Chromosome abnormalities defined by conventional cytogenetics in plasma cell leukemia: what have we learned about its biology? Eur J Haematol. 2011; 87:20–27. PMID: 21692850.

19. Oh S, Koo DH, Kwon MJ, et al. Chromosome 13 deletion and hypodiploidy on conventional cytogenetics are robust prognostic factors in Korean multiple myeloma patients: web-based multicenter registry study. Ann Hematol. 2014; 93:1353–1361. PMID: 24671365.

20. Caltagirone S, Ruggeri M, Aschero S, et al. Chromosome 1 abnormalities in elderly patients with newly diagnosed multiple myeloma treated with novel therapies. Haematologica. 2014; 99:1611–1617. PMID: 25015938.

21. An G, Xu Y, Shi L, et al. Chromosome 1q21 gains confer inferior outcomes in multiple myeloma treated with bortezomib but copy number variation and percentage of plasma cells involved have no additional prognostic value. Haematologica. 2014; 99:353–359. PMID: 24213147.

22. Hebraud B, Magrangeas F, Cleynen A, et al. Role of additional chromosomal changes in the prognostic value of t(4;14) and del(17p) in multiple myeloma: the IFM experience. Blood. 2015; 125:2095–2100. PMID: 25636340.

23. Chretien ML, Corre J, Lauwers-Cances V, et al. Understanding the role of hyperdiploidy in myeloma prognosis: which trisomies really matter? Blood. 2015; 126:2713–2719. PMID: 26516228.

Fig. 1
The number of structural aberrations detected on each chromosome in patients with multiple myeloma. Structural abnormalities involving chromosome 1 were the most common, followed by aberrations of chromosomes 14, 11, and 8.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download