Dear Editor,
Bordetella, of the family Alcaligenaceae, is a genus containing gram-negative coccoid rod species. To date, nine species have been assigned to the genus Bordetella [1]. Human infections caused by B. petrii have seldom been reported since its first detection in 2001 [2345678]. Among seven reported cases, four were recovered from respiratory specimens [4568], while three were recovered from pus specimens of patients with bone-related infections [237]. A 52-yr-old man without any medical history was admitted to a tertiary-care hospital (Hospital A) in Seoul, Korea, for tibia and fibula fractures of both legs. He had sustained a crush injury to both legs on a riverboat in Thailand. He had undergone surgery and subsequently presented with purulent discharge from the muscle flap site of the left leg. The wound culture revealed a gram-negative rod identified as Achromobacter denitrificans by using a VITEK 2 GNI card (bioMérieux, Marcy l'Etoile, France). After a month of managing the abscess site, the patient was transferred to another hospital (Hospital B) in Ilsan, Korea. At that hospital, deep tissue and bone cultures revealed the presence of Bordetella bronchiseptica and Alcaligenes species, respectively, using MicroScan (Dade Behring, West Sacramento, CA, USA). During a 15-month follow-up, nonunion of fractured sites was observed. The patient was then admitted to our hospital (Hospital C) in Seoul, Korea, to undergo surgery for the delayed union. After discharge, his leg wound was managed at an outpatient clinic because his laboratory tests carried out three days after the surgery showed leukocytosis (14×109/L) with elevated C-reactive protein (CRP) level (182.2 mg/L).
Deep wound culture yielded medium to large grayish and convex colonies that were non-hemolytic on sheep blood agar plate (BAP), and colorless, opaque pinpoint-sized colonies on MacConkey agar plate (isolate C1). Two serial follow-up cultures revealed the same species (isolates C2 and C3). Smear preparation of these colonies showed short gram-negative rods. Subculture on triple-sugar iron slant agar showed that the bacteria did not ferment glucose or produce H2S. Isolates exhibited an umbrella-shaped growth pattern when cultured in motility-indole-ornithine semisolid agar, suggesting that the microorganism is strictly aerophilic and motile. API 20NE (bioMérieux) and VITEK 2 GNI cards identified the isolates as A. denitrificans and Burkholderia species, respectively. However, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS; Bruker Daltonics, Billerica, MA, USA) identified them as B. petrii with a 2.089 score. The isolates were finally confirmed as B. petrii on the basis of partial sequences of the 16S rRNA gene, which showed 100% identity to those of DSM 12804, the type strain of B. petrii, when analyzed by BLASTn (http://www.ncbi.nlm.nih.gov/BLAST). XbaI-macrorestriction analysis was performed by using pulse field gel electrophoresis (PFGE) (CHEF-DRII System, Bio-Rad, Hercules, CA, USA), as described previously [9]. All three clinical isolates exhibited identical PFGE banding patterns (Fig. 1).
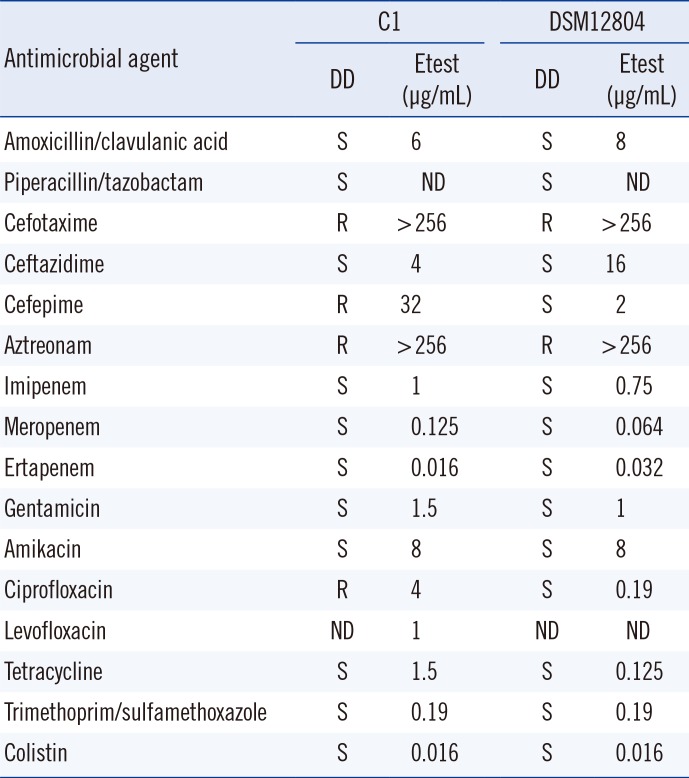
Antimicrobial susceptibility testing (AST) was performed with the disk diffusion method. The minimal inhibition concentrations (MICs) of antimicrobials were determined by Etest according to the Clinical and Laboratory Standard Institute guidelines [10]. Clinical isolates C1-3 exhibited antimicrobial susceptibility patterns similar to those of the type strain DSM 12804. However, all three clinical isolates were resistant to cefepime and ciprofloxacin, while the type strain was susceptible to both drugs (Table 1). Cephalexin was used as empirical therapy and changed to levofloxacin following AST results. After two months of treatment, the wound infection was resolved with the normalization of the white blood cell (WBC) count (9.22×109/L) and CRP level (3.3 mg/L). Isolates C1-3 may be clones because they exhibited identical antimicrobial susceptibility and PFGE banding patterns. Former clinical isolates were identified as A. denitrificans, B. bronchiseptica, and Alcaligenes species in hospitals A and B. However, all these isolates are speculated to be B. petrii because commercially available cards misidentified our three clinical isolates and type strain DSM 12804 as A. denitrificans and Burkholderia species. Discrepancies among identification methods have been reported previously [35]. However, MALDI-TOF MS identified the isolates correctly with a high score. When bacterial culture results show discrepancies among different bacterial identification methods and the results include members of Alcaligenaceae, B. petrii should be suspected and confirmed by using MALDI-TOF MS or 16S rRNA sequencing.
Here, B. petrii infection persisted for more than one year. Long-lasting B. petrii infections have been reported previously [5]. Chronic infections, despite continuous use of antibiotics, suggest the clinical difficulty of treating B. petrii. Bone-related infections are associated with reduced effects of antimicrobial therapy, which possibly contribute to persistent B. petrii infection.
Acknowledgments
This work was supported by a grant from the Research Program funded by the Korea Center for Disease Control and Prevention (#2014E4700201).
References
1. Ko KS, Peck KR, Oh WS, Lee NY, Lee JH, Song JH. New species of Bordetella, Bordetella ansorpii sp. nov., isolated from the purulent exudate of an epidermal cyst. J Clin Microbiol. 2005; 43:2516–2519. PMID: 15872300.
2. Fry NK, Duncan J, Malnick H, Warner M, Smith AJ, Jackson MS, et al. Bordetella petrii clinical isolate. Emerg Infect Dis. 2005; 11:1131–1133. PMID: 16022798.
3. Stark D, Riley LA, Harkness J, Marriott D. Bordetella petrii from a clinical sample in Australia: isolation and molecular identification. J Med Microbiol. 2007; 56:435–437. PMID: 17314377.
4. Spilker T, Liwienski AA, LiPuma JJ. Identification of Bordetella spp. in respiratory specimens from individuals with cystic fibrosis. Clin Microbiol Infect. 2008; 14:504–506. PMID: 18325036.
5. Le Coustumier A, Njamkepo E, Cattoir V, Guillot S, Guiso N. Bordetella petrii infection with long-lasting persistence in human. Emerg Infect Dis. 2011; 17:612–618. PMID: 21470449.
6. Zelazny AM, Ding L, Goldberg JB, Mijares LA, Conlan S, Conville PS, et al. Adaptability and persistence of the emerging pathogen Bordetella petrii. PLoS One. 2013; 8:e65102. PMID: 23750235.
7. Nogi M, Bankowski MJ, Pien FD. Septic arthritis and osteomyelitis due to Bordetella petrii. J Clin Microbiol. 2015; 53:1024–1027. PMID: 25540393.
8. Nagata JM, Charville GW, Klotz JM, Wickremasinghe WR, Kann DC, Schwenk HT, et al. Bordetella petrii sinusitis in an immunocompromised adolescent. Pediatr Infect Dis J. 2015; 34:458. PMID: 25760569.
9. Mooi FR, Hallander H, Wirsing von König CH, Hoet B, Guiso N. Epidemiological typing of Bordetella pertussis isolates: recommendations for a standard methodology. Eur J Clin Microbiol Infect Dis. 2000; 19:174–181. PMID: 10795589.
10. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 24th Informational supplement, M100-S24. Wayne, PA: Clinical and Laboratory Standards Institute;2014.
Fig. 1
Pulsed field gel electrophoresis (PFGE) was performed by using the CHEF-DRII System (Bio-Rad, Hercules, CA, USA) at 6 V/cm for 20 hr at 10℃, with initial and final pulse times of 0.5 sec and 30 sec, respectively. XbaI was the restriction enzyme used. All three clinical isolates from our hospital (hospital C) exhibited 100% identical PFGE banding patterns. This finding suggests that sequentially identified B. petrii were the same strains. Lane C1-3, Bordetella petrii isolated from patient's wound; ATCC, ATCC DSM 12804.
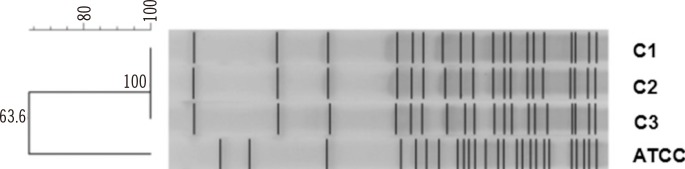
Table 1
Antimicrobial susceptibility profiles




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download