Dear Editor,
The Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer (MDBGFC) has more than 100 cases for the t(9;22)(q34;q11.2) with divergent patterns of clonal evolution [1]. Here, we present a patient with B-cell ALL who showed t(9;22)(q34;q11.2) and a clonal divergence.
Hematological studies from a 54-yr-old woman revealed white blood cell count of 97.2×109/L, platelets 35×109/L, and hemoglobin 6.9 g/dL. Bone marrow blasts displayed L2 morphology and expressed cell-surface differentiation antigens defining a B-cell phenotype, namely, CD10 (80%), CD19 (78%), CD5 (11%), CD3 (0%), and CD13 (0%). The patient received vincristine, prednisone, and daunorubicin-based chemotherapy. However, four months later, the patient was in the terminal phase and died after infiltrations were detected both to the retro-ocular and central nervous systems.
We analyzed metaphasic cells from non-stimulated peripheral blood samples obtained eight days before the patient's death. Her karyotype included several sub-clones, which were described according to the International System for Human Cytogenetic Nomenclature (ISCN) (2013) [2], as: 46,XX,t(9;22)(q34;q11.2)[3]/46,idem,ider(22)(22pter→22q11.2::9q34→9q?tel::9q?tel→9q34::22q11.2→22pter)[14]/46,idem,t(13;17)(q14;q25)[3]/46,idem,+1,dic(1;1)(?;?),t(13;17)(q14;q25)[8]/46,XX[1] (Fig. 1A-E). C-banding by barium hydroxide and Giemsa revealed two heterochromatic regions (1q12 band) on the dic(1;1) chromosome (Fig. 1F).
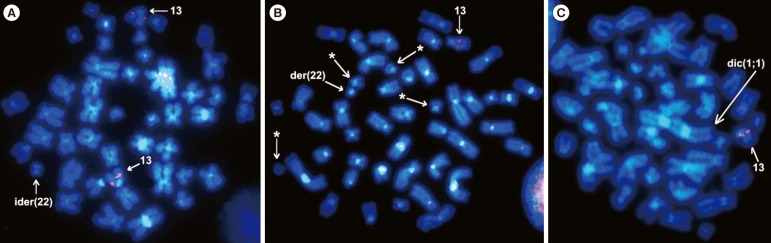
FISH studies with the dual-color, single-fusion BCR/ABL1 probe (Vysis, Downers Grove, IL, USA) revealed two fusion signals on the ider(22) and only one fusion signal on the standard Philadelphia chromosome (data not shown). FISH study with the RB1 probe (Vysis) disclosed two signals on both normal 13 chromosomes in the clone containing ider(22) (Fig. 2A). Cells with t(13;17) were positive only on the normal chromosome 13 and did not display any signal on either der(13) or der(17) (Fig. 2B and C), thus demonstrating the loss of RB1 by translocation.
Yamamoto et al. [3] reported the presence of first and only ider(22) chromosome similar to the one observed by us in another ALL patient who had a poor prognosis too. These ider(22) chromosomes could result from an U-type sister chromatid exchange. The ider(22) chromosome equals a second copy of der(22) chromosome, which is well known to be one of the main secondary chromosomal changes related to the clonal evolution of cells with t(9;22) [1].
The co-occurrence of t(9;22)(q34;q11) and t(13q14) is rare. There are only six cases registered in the MDBGFC [1], which were previously reported [4567]. When both translocations were present, the patients had CML in blast crisis or relapse. Out of four patients tested for chromosome 13 deletions by using FISH [56], three tested negative, and only in the fourth patient, there was inconclusive evidence of such a deletion [6].
The t(13;17)(q14;q25), which caused the loss of RB1 sequences in the present patient has not been previously described in ALL [1]. This translocation has only been detected in an AML-M4 patient who did not show deletion of the RB1 gene [8], and in a case of Sézary syndrome in which the status of RB1 gene was not tested [9]. Although we did not know the extent of present 13q deletion, some features of the patient such as genome instability, poor prognosis, and shortened survival suggest that she had a large deletion inclusive of the RB1 gene. Moreover, the chromosomal band 17q25 has been implicated in several translocations also associated with poor prognosis and shorter survival [1].
Imbalances in chromosome 1 are frequent in human neoplasia [1]. The apparently different sizes of two heterochromatic regions observed on the dic (1;1) chromosome (Fig. 1F and 2C) suggest a translocation between homologous chromosomes as the forming mechanism of such a dicentric chromosome.
Concomitant with the clonal divergence found in peripheral blood, this patient had central nervous system and retro-ocular infiltrations that could not be further studied. Nowicki et al. [10] observed that the circulating CML-blast crisis cells showed major changes than the CML-cells from bone marrow. This observation suggests that leukemic bloodstream cells differ from those in their original niche in survival, growth, invasiveness, and metastatic potential. In our patient, we could not determine whether the divergent sub-clones found in peripheral blood were also present in the infiltrations, or whether each infiltration was derived from a unique and independent sub-clone. This case calls for research on chromosomal and genetic changes that enable the leukemic cells to infiltrate other tissues.
Acknowledgements
We thank Dr. Horacio Rivera for his academic criticism and María de Lourdes Carbajal for reviewing our manuscript. This work was funded by the CONACyT # SALUD-2005-C01-13870 and by the FIS-IMSS # FIS/IMSS/PROT/G12/1138.
References
1. Mitelman F, Johansson B, Mertens F, editors. Mitelman database of chromosome aberrations and gene fusions in cancer. Updated on Nov 19, 2014. http://cgap.nci.nih.gov/Chromosomes/Mitelman.
2. Shaffer LG, McGowan-Jordan J, Schmid M, editors. ISCN (2013): An International System for Human Cytogenetic Nomenclature. Basel: S. Karger;2013.
3. Yamamoto K, Nagata K, Morita Y, Inagaki K, Hamaguchi H. Isodicentric Philadelphia chromosome in acute lymphoblastic leukemia with der (7;12)(q10;q10). Leuk Res. 2007; 31:713–718. PMID: 16979235.
4. Carbone P, Granata G, Margiotta G, Barbata G, Majolino I. Ph1 duplication, t(13q-; 14q+) and trisomy 19 in a case with chronic myeloid leukemia in lymphoid blast crisis at presentation. Haematologica. 1982; 67:595–604. PMID: 6815018.
5. Coignet LJ, Lima CS, Min T, Streubel B, Swansbury J, Telford N, et al. Myeloid- and lymphoid-specific breakpoint cluster regions in chromosome band 13q14 in acute leukemia. Genes Chromosomes Cancer. 1999; 25:222–229. PMID: 10379868.

6. Chase A, Pickard J, Szydlo R, Coulthard S, Goldman JM, Cross NC. Non-random involvement of chromosome 13 in patients with persistent or relapsed disease after bone-marrow transplantation for chronic myeloid leukemia. Genes Chromosomes Cancer. 2000; 27:278–284. PMID: 10679917.

7. Kim YJ, Kim DW, Lee S, Kim YL, Hwang JY, Park YH, et al. Cytogenetic clonal evolution alone in CML relapse post-transplantation does not adversely affect response to imatinib mesylate treatment. Bone Marrow Transplant. 2004; 33:237–242. PMID: 14628081.

8. Turhan N, Yürür-Kutlay N, Topcuoglu P, Sayki M, Yüskel M, Grman G, et al. Translocation (13;17)(q14;q25) as a novel chromosomal abnormality in acute myeloid leukemia-M4. Leuk Res. 2006; 30:903–905. PMID: 16469377.

9. Johnson GA, Dewald GW, Strand WR, Winkelmann RK. Chromosome studies in 17 patients with the Sézary syndrome. Cancer. 1985; 55:2426–2433. PMID: 3857106.

10. Nowicki MO, Pawlowski P, Fischer T, Hess G, Pawlowski T, Skorski T. Chronic myelogenous leukemia molecular signature. Oncogene. 2003; 22:3952–3963. PMID: 12813469.

Fig. 1
Chromosomes banded with standard techniques. (A) Metaphasic cell with the ider(22). (B) This metaphasic cell contains both t(9;22)(q34;q11.2) and t(13;17)(q14;q25). (C) In addition to the two translocations observed in the above image, a dic(1;1) chromosome is shown whose breakpoints could not be ascertained. In this picture, a single-cell chromosomal translocation t(X;15) is seen; however, it was not included in the karyotype formula based on the International System for Human Cytogenetic Nomenclature (2013) criteria [2]. Selected chromosomes: (D) 22 and ider(22); and, (E) 1 plus dic(1;1). (F) Selected C--banded by barium hydroxide and Giemsa (CBG) chromosomes 1 and dic(1;1). Note that the heterochromatic regions on the dic(1;1) chromosome apparently have different sizes, the bigger heterochromatic region being more similar to the one present in the normal chromosome 1 (×100 for A, B, and C).
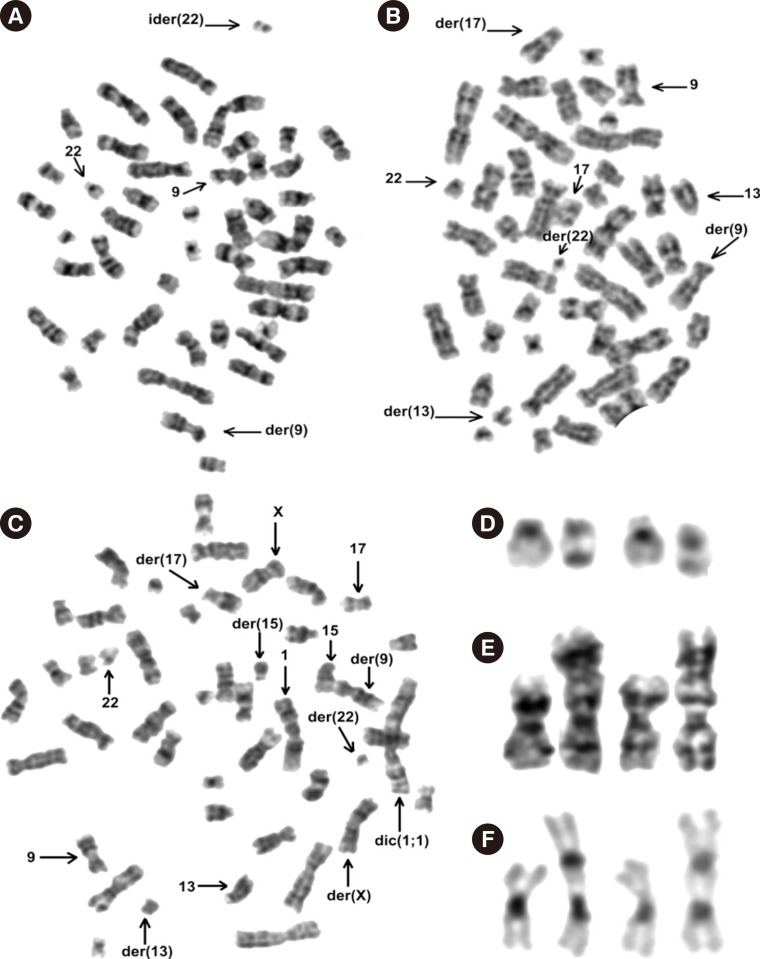
Fig. 2
FISH analysis with the RB1 probe (red signals) in cells counterstained with DAPI (4',6-diamidino-2-phenylindole; blue color). (A) Metaphase from the clone with ider(22) showing two RB1 signals on both normal 13 chromosomes. (B) Cell with t(13;17) displaying only one RB1-positive signal on the normal chromosome 13; asterisks show four G group chromosomes one of which is der(13). (C) Cell with t(13;17) plus dic(1;1) also shows only one RB1-positive signal on the normal chromosome 13 (×100).




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download