Abstract
Background
We evaluated the coincidence rate between Vitek MS system (bioMérieux, France) and Vitek 2 in identifying uropathogens directly from urine specimens.
Methods
Urine specimens submitted to our microbiology laboratory between July and September 2013 for Gram staining and bacterial culture were analyzed. Bacterial identification was performed by using the conventional method. Urine specimens showing a single morphotype by Gram staining were processed by culturing and matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). Of 2,370 urine specimens, 251 showed a single morphotype on Gram staining, and among them, 202 were available for MALDI-TOF MS.
Results
In these 202 specimens, colony growth was observed in 189 specimens, and 145 specimens had significant growth of single-colony morphotype in culture. One hundred and ten (75.9%) of them had colony counts of ≥105 colony-forming units (CFU)/mL and included 71 enteric gram-negative bacteria (GNB), 5 glucose-non-fermenting GNB, 9 gram-positive cocci (GPC), and 25 yeasts. Furthermore, 70 (98.6%), 3 (60.0%), 4 (44.4%), and 5 (20.0%), respectively, of these were correctly identified by Vitek MS. Thirty-one specimens (21.4%; 11 GNB, 7 GPC, 12 yeasts, and 1 gram-positive bacillus) had colony counts of 104-105 CFU/mL. Four specimens (2.8%) yielded colony counts of 103-104 CFU/mL.
Conclusions
Vitek MS showed high rate of accuracy for the identification of GNB in urine specimens (≥105 CFU/mL). This could become a rapid and accurate diagnostic method for urinary tract infection caused by GNB. However, for the identification of GPC and yeasts, further studies on appropriate pre-treatment are warranted.
Urinary tract infection (UTI) is the most commonly encountered nosocomial infection, and the major risk factor is urinary catheterization [1]. Although Escherichia coli is the most common uropathogen for UTI, other gram-negative bacteria (GNB; enterobacteria other than E. coli, Pseudomonas spp., and Acinetobacter spp.), gram-positive cocci (GPC), and Candida spp. are common uropathogens. In patients positive for the same microorganism in both urine and blood culture, the five leading microorganisms found in blood cultures are E. coli, Candida spp., Klebsiella spp., Acinetobacter spp., and Staphylococcus aureus [2]. Therefore, early identification of uropathogens is necessary, especially in nosocomial UTI, and it could help select appropriate antibiotics for treatment. However, urine culture (quantitative culture of urine specimens on solid medium followed by biochemical characterization of isolates), which is the gold standard for diagnosis, takes 24-72 hr before results are available [3].
Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) becomes a reliable tool for the identification of bacteria and yeast [4, 5]. The Vitek MS system (bioMérieux, Marcy I'Etoile, France) got US Food and Drug Administration (FDA) clearance on August 2013 for the identification of bacteria (except Mycobacterium) and yeasts grown on solid media. It was recently reported that MALDI-TOF MS could directly provide rapid and accurate bacterial identification for the majority of urine specimens with a bacterial colony count of ≥105 colony-forming units (CFU)/mL, where an automated device based on flow cytometry was used for the screening test [6, 7]. However, in most clinical laboratories, such an automated device is not used for screening UTIs. For urine specimens with ≥104 CFU/mL of a single uropathogen, the identification and susceptibility test for the uropathogen is recommended because colony counts of <105 CFU/mL in voided specimens in the presence of dysuria and symptoms of UTI should not be ignored [8]. In this study, we evaluated the coincidence rate between Vitek MS system (bioMérieux, France) and Vitek 2 in identifying urinary tract pathogens directly from urine specimens with ≥105 CFU/mL, as well as those with 104-105 CFU/mL.
Of the urine specimens submitted to our microbiology laboratory between July and September 2013 for Gram staining and bacterial culture, specimens showing a single morphotype by Gram staining were processed both by conventional culture and MALDI-TOF MS. Leukocyte numbers were examined at a low-power field (LPF) and were graded as negative, 1+:<1/LPF, 2+: 1-9/LPF, 3+:10-25/LPF, or 4+:>25/LPF. Bacteria were counted per oil immersion field (OIF) and graded as negative, 1+:<1/OIF, 2+:1-5/OIF, 3+:6-30/OIF, or 4+:>30/OIF [8].
Bacterial identification was performed by the conventional method using the Vitek 2 system. For the conventional culture, 1 µL of well-mixed urine was inoculated and spread onto blood agar plates and MacConkey agar plates using a sterile plastic disposable loop (SPL Lifesciences, Pocheon, Korea). Plates were incubated in an aerobic atmosphere at 37℃ for 18-24 hr. When bacterial growth was observed, the colonies on blood agar were counted, and colonies from both types of plates were identified by using the Vitek 2 system.
For MALDI-TOF MS analysis, the preparation method was an adaptation of the protocol described by Ferreira et al. [7] and Lavergne et al. [9]. Briefly, urine (3 mL) was centrifuged at 2,000 g for 30 sec to remove leukocytes. The supernatant was centrifuged at 14,100 g for 10 min to collect bacteria. The pellet was washed once with de-ionized water (DW). To the pellet, 1.5 mL of basic solution of sodium dodecyl sulfate (SDS 0.1%, NaHCO3 0.015M) was added and incubated at 37℃ for 10 min. After further centrifugation at 14,100 g for 10 min and washing with DW, the supernatant was removed. The pellet itself (8-10 µL) was applied to the MALDI plate in duplicate and was dried on a slide warmer (56℃) for 10 min. Then, 1 µL of matrix solution (3.1% [w/v] α-cyano-4-hydroxycinnamic acid, product no. 411071; bioMérieux) was added to each well. For yeasts, 1 µL of formic acid (product no. 411072, bioMérieux) was applied before the addition of matrix solution. No repeat testing was performed.
Following preparation, specimens were analyzed with the Vitek MS in linear, positive-ion mode at a laser frequency of 200 Hz, across the mass-to-charge (m/z) ratio of 2,000 to 20,000 Da. External mass calibration was performed by using a TOF mix (bioMérieux). Target plates were calibrated and quality controlled using E. coli ATCC8739. After the acquisition of spectra, data were transferred from the Vitek MS acquisition station to the Vitek MS analysis server, and identification results were displayed by using Myla v3.2 middleware. The Vitek MS identification system is based on comparison of the characteristics of the obtained spectra with those of the Vitek MS v2.0 database. This database was built by using spectra of known strains for each claimed species. On the basis of this representative data collection, a weight is assigned to each peak for each species according to its specificity.
A confidence value of ≥60 with the unique spectrum of a single organism indicated good species-level identification. If no unique identification pattern was found, a list of possible organisms was given as "low discrimination" (confidence value of <60%) or the strain was determined to be outside the scope of the database ("no ID") [4]. If one of the duplicates showed a confidence value of ≥60%, it was considered an acceptable identification. No identification result was given when the duplicate had poor confidence levels or did not have the same species-level identification. The warning messages "bad spectrum" or "insufficient peak" appeared in case of a human error or a poor-quality deposit.
Statistical analyses were performed by using the SPSS statistical analysis package version 13.0 (SPSS Inc., Chicago, IL, USA). Comparisons of proportions were tested by using chi-square analysis and Fisher's exact test, as appropriate, and the criterion for statistical significance was set at P<0.05.
Of the 202 urine specimens, the growth of colonies was observed in 189 specimens: 145 specimens with single-colony morphology, 22 specimens with two-colony morphology, and the remaining 22 specimens with morphologies of more than two colonies. Thirteen specimens did not grow in culture, and Vitek MS did not identify any significant protein profile in any of these cases (Table 1).
Of the 145 specimens with significant growth of single-colony morphology on agar, 110 (75.9%) of them had colony counts of ≥105 CFU/mL. They included 71 enteric GNB, five glucose-non-fermenting GNB, nine GPC, and 25 yeasts. Vitek MS correctly identified 70 (98.6%) enteric GNB, three (60.0%) glucose-non-fermenting GNB, four (44.4%) GPC, and five (20.0%) yeast isolates (Table 2). Overall, among the 110 specimens with colony counts of ≥105 CFU/mL, microorganism identification coincided at the species level in 79 cases (71.8%) and at the genus level in 84 cases (76.4%). In 26 cases, Vitek MS did not give a reliable identification. We found no major discrepancies at the genus level.
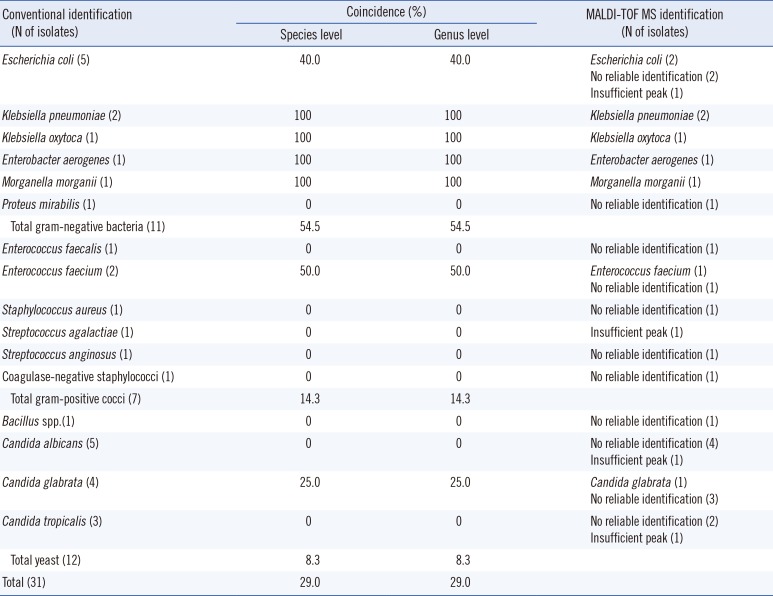
Thirty-one specimens (31/145, 21.4%) showed growth of bacterial colonies between 104 and 105 CFU/mL, and they were 11 GNB, seven GPC, 12 yeasts, and one gram-positive bacillus (GPB) (Table 3). Among them, seven (63.6%) GNB, one (14.3%) GPC, and one (8.3%) yeast were correctly identified by Vitek MS. Overall, the accuracy of identification at the genus level was 29.0% (9/31). The correct identification rate was significantly higher in urine specimens with ≥105 CFU/mL than in those with 104-105 CFU/mL (P=0.000).
Four specimens (2.8%) yielded colony counts between 103 and 104 CFU/mL, and none of them were correctly identified.
According to Gram staining of the microorganisms isolated from the above 141 specimens from which more than 104 CFU/mL of an microorganism was isolated, 87 were GNB, 16 were GPC, 37 were yeasts, and one was GPB. Of the 87 GNB cases, E. coli was isolated in 58 (66.7%), Klebsiella spp. in 15 (17.2%), chromosomal AmpC-producing Enterobacteriaceae (Enterobacter spp., Citrobacter spp., Serratia spp., Morganella spp.) in seven (8.0%), glucose non-fermenting isolates in five (5.7%), and Proteus spp. in two cases. Overall, the coincidence rate between Vitek 2 and Vitek MS was 92.0% (80/87) for GNB (54/58 E. coli, 15/15 Klebsiella spp., 7/7 chromosomal AmpC-producing Enterobacteriaceae, 3/5 glucose non-fermenting GNB, and 1/2 Proteus spp.), 31.3% (5/16) for GPC (3/6 Enterococcus faecalis, 1/4 Enterococcus faecium, 1/3 Streptococcus agalactiae, 0/1 Staphylococcus aureus, 0/1 Streptococcus anginosus group, 0/1 coagulase-negative Staphylococcus), 16.2% (6/37) for yeasts, and 0% for GPB (0/1 Bacillus spp.). Among the 50 specimens not identified by Vitek MS, eight had "insufficient peaks" caused by poor quality of the microbial film on the plate, and five of them were yeasts. The number of specimens with high or moderate leukocytes were 28 among the 91 cases correctly identified by Vitek MS, and six among the 50 cases not identified by Vitek MS (chi-square test, P=0.003).
For 22 specimens with significant (≥104 CFU/mL) growth of two-colony morphology in culture, Vitek MS correctly identified one of the mixed microorganisms in seven cases (two E. coli, two Acinetobacter baumannii complex, one Klebsiella pneumoniae, one Morganella morganii, and one Streptococcus agalactiae).
Previous studies have shown excellent correlation between Vitek MS identification and conventional microbiological identification in clinical bacterial and yeast isolates. In these studies, when there were discrepancies between Vitek MS and conventional identification, sequencing results frequently corresponded with the Vitek MS [4, 5, 10]. However, few studies on direct identification of urinary tract pathogens from urine specimens using MALDI-TOF MS have been performed [6, 7], and to our knowledge, this is the first such study that used Vitek MS and included specimens with yeasts.
Among urine specimens containing ≥104 CFU/mL, GNB accounted for 61.7% of the uropathogens, and yeast was the second most common (26.2%). Moreover, among the GNB, E. coli comprised only about two thirds, which is in line with a previous study from China where E. coli was found to comprise 43.5% of the uropathogens. Considering this diversity of uropathogens, early identification of uropathogens is necessary for appropriate therapy [6].
Based on our results, the overall identification rate of Vitek MS was much higher (76.4%) in urine specimens containing ≥105 CFU/mL than that in the specimens containing 104-105 CFU/mL (29.0%). For GPC and yeasts, the identification rate in specimens with a lower microbial burden was too low (14.3% and 8.3%, respectively). However, for GNB, the identification rate was 63.6% even for specimens with 104 or 105 CFU/mL of bacteria, which is much higher than that reported in an earlier study where only one of the five GNB were correctly identified [7]. For urine specimens containing bacteria of less than 104 CFU/mL, Vitek MS could not identify bacteria. This indicates that sufficient bacterial count is essential for identification by MALDI-TOF MS and supports the previous report where bacterial count needed for a good discrimination score by MALDI-TOF MS was higher for GPC (2.5-5×105 CFU/mL) than for GNB (about 6×104 CFU/mL) [6]. Therefore, additional studies are needed to improve the collection quantity and purity of collected bacteria.
For urine specimens with ≥105 CFU/mL, the identification rate differed according to the microorganisms involved. For enteric GNB, Vitek MS demonstrated excellent identification (98.6%), and this finding is in line with previous studies [6, 7]. For GPC, however, Vitek MS yielded correct identification only in a small portion (44.4%) of urine specimens with GPC. For E. faecalis, which is the most common uropathogenic GPC, although the number of specimens was small, the detection rate was 60%, which corresponds with a study where the coincidence with conventional culture method was lower (66.7%) in E. faecalis than in E. coli (97.6%) [7]. However, in the study from China, the coincidence was very high in Enterococcus spp. (81.5-90.0%) [6]. This difference might be attributed to the difference in specimen preparation, because those authors used an extraction procedure using formic acid and acetonitrile [6], while we did not (to avoid the use of a fume hood). Perhaps the formic acid and acetonitrile extraction procedure would have a positive effect on the identification of GPC by MALDI-TOF MS. For specimens containing yeasts, only five (20%) out of the 25 specimens were correctly identified. As Vitek MS showed superior ability of identification of yeasts grown on agar (86.7-96.6%) using the simple formic acid pre-treatment [5, 8, 11, 12], the low identification rate for yeast in this study can be ascribed to the matrix effect of urine. A better pre-treatment step that will allow for high identification rate for yeasts directly from urine specimens needs to be developed.
We also compared leukocyte counts between specimens that were identified or not identified by Vitek MS, because in a previous study [6], a large number of leukocytes in urine specimens was frequently observed in specimens not reliably identified by MALDI-TOF MS. However, the proportion of urine specimens containing a large number of leukocytes was much higher among the specimens correctly identified by Vitek MS, suggesting that a high leukocyte count does not interfere with bacterial protein peaks. High concentrates of many endogenous substances such as α-defensins in urine, which can suppress or enhance the ionization of certain protein spectra of microorganisms, might result in incorrect or no identification [13, 14].
In our study, the identification of bacteria by MALDI-TOF MS took about 1 hr, which is similar to that reported in several studies [15, 16]. Identifying bacterial species will help clinicians choose appropriate antibiotics for empiric therapy. In addition, when the bacterial species is known, the antimicrobial susceptibility test can be performed as soon as the colony is formed on the agar, using the appropriate Vitek card (AST-N224 for Enterobacteriaceae vs. AST-N225 for glucose non-fermenters, AST-P600 for Enterococcus spp.).
In conclusion, with a simple pre-treatment using SDS, Vitek MS showed high rate of accuracy for the identification of GNB in urine specimens with ≥104 CFU/mL, without any major error. This could become a rapid and accurate diagnostic method useful in the diagnosis of UTI caused by a single GNB. However, for the identification of GPC and yeasts, further studies are required to improve pre-treatment steps. In addition, Vitek MS could not identify bacterial species in urine specimens with mixed bacterial infections; therefore, for improvements in the Vitek MS software, this shortcoming should be considered and rectified.
Acknowledgments
This work was supported by bioMérieux Korea, but they were not involved in the data collection or preparation of the manuscript.
References
1. Trautner BW. Management of catheter-associated urinary tract infection. Curr Opin Infect Dis. 2010; 23:76–82. PMID: 19926986.

2. Isikgoz Tasbakan M, Durusoy R, Pullukcu H, Sipahi OR, Ulusoy S. 2011 Turkish Nosocomial Urinary Tract Infection Study Group. Hospital-acquired urinary tract infection point prevalence in Turkey: differences in risk factors among patient groups. Ann Clin Microbiol Antimicrob. 2013; 12:31. PMID: 24188193.

3. Burd EM. A critical appraisal of the role of the clinical microbiology laboratory in the diagnosis of urinary tract infections. J Clin Microbiol. 2011; 49(S9):S34–S38.

4. Dubois D, Grare M, Prere MF, Segonds C, Marty N, Oswald E. Performances of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for rapid identification of bacteria in routine clinical microbiology. J Clin Microbiol. 2012; 50:2568–2576. PMID: 22593596.

5. Won EJ, Shin JH, Lee K, Kim MN, Lee HS, Park YJ, et al. Accuracy of species-level identification of yeast isolates from blood cultures from 10 university hospitals in South Korea by use of the matrix-assisted laser desorption ionization-time of flight mass spectrometry-based Vitek MS system. J Clin Microbiol. 2013; 51:3063–3065. PMID: 23784123.

6. Wang XH, Zhang G, Fan YY, Yang X, Sui WJ, Lu XX. Direct identification of bacteria causing urinary tract infections by combining matrix-assisted laser desorption ionization-time of flight mass spectrometry with UF-1000i urine flow cytometry. J Microbiol Methods. 2013; 92:231–235. PMID: 23305925.

7. Ferreira L, Sánchez-Juanes F, González-Avila M, Cembrero-Fuciños D, Herrero-Hernández A, González-Buitrago JM, et al. Direct identification of urinary tract pathogens from urine samples by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2010; 48:2110–2115. PMID: 20392910.

8. Garcia LS, editor. Clinical microbiology procedure handbook. 3rd ed. Washington DC: ASM press;2010. p. 3.2.16.
9. Lavergne RA, Chauvin P, Valentin A, Fillaux J, Roques-Malecaze C, Arnaud S, et al. An extraction method of positive blood cultures for direct identification of Candida species by Vitek MS matrix-assisted laser desorption ionization time of flight mass spectrometry. Med Mycol. 2013; 51:652–656. PMID: 23373445.
10. Moon HW, Lee SH, Chung HS, Lee M, Lee K. Performance of the Vitek MS matrix-assisted laser desorption ionization time-of-flight mass spectrometry system for identification of Gram-positive cocci routinely isolated in clinical microbiology laboratories. J Med Microbiol. 2013; 62:1301–1306. PMID: 23764744.

11. Jamal WY, Ahmad S, Khan ZU, Rotimi VO. Comparative evaluation of two matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems for the identification of clinically significant yeasts. Int J Infect Dis. 2014; 26:167–170. PMID: 25080355.

12. Westblade LF, Jennemann R, Branda JA, Bythrow M, Ferraro MJ, Garner OB, et al. Multicenter study evaluating the Vitek MS system for identification of medically important yeasts. J Clin Microbiol. 2013; 51:2267–2272. PMID: 23658267.

13. Tudela E, Muñoz G, Muñoz-Guerra JA. Matrix effect marker for multianalyte analysis by LC-MS/MS in biological samples. J Chromatogr B Analyt Technol Biomed Life Sci. 2012; 901:98–106.

14. Köhling HL, Bittner A, Müller KD, Buer J, Becker M, Rübben H, et al. Direct identification of bacteria in urine samples by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and relevance of defensins as interfering factors. J Med Microbiol. 2012; 61:339–344. PMID: 22275503.

15. Drancourt M. Detection of microorganisms in blood specimens using matrix-assisted laser desorption ionization time-of-flight mass spectrometry: a review. Clin Microbiol Infect. 2010; 16:1620–1625. PMID: 20545958.

16. Guo L, Ye L, Zhao Q, Ma Y, Yang J, Luo Y. Comparative study of MALDI-TOF MS and VITEK 2 in bacteria identification. J Thorac Dis. 2014; 6:534–538. PMID: 24822115.
Table 1
Urine culture results of 202 specimens available for MALDI-TOF MS

| Urine culture | N of case |
|---|---|
| Growth of colonies (N = 189) | |
| 1-colony morphology | 145 |
| ≥ 105 CFU/mL | 110 |
| 104-105 CFU/mL | 31 |
| 103-104 CFU/mL | 4 |
| 2-colony morphology | 22 |
| 3-colony morphology | 22 |
| No growth (N = 13) |
Table 2
Comparison of the results between the conventional method and MALDI-TOF in 110 specimens with colony counts of ≥105 CFU/mL
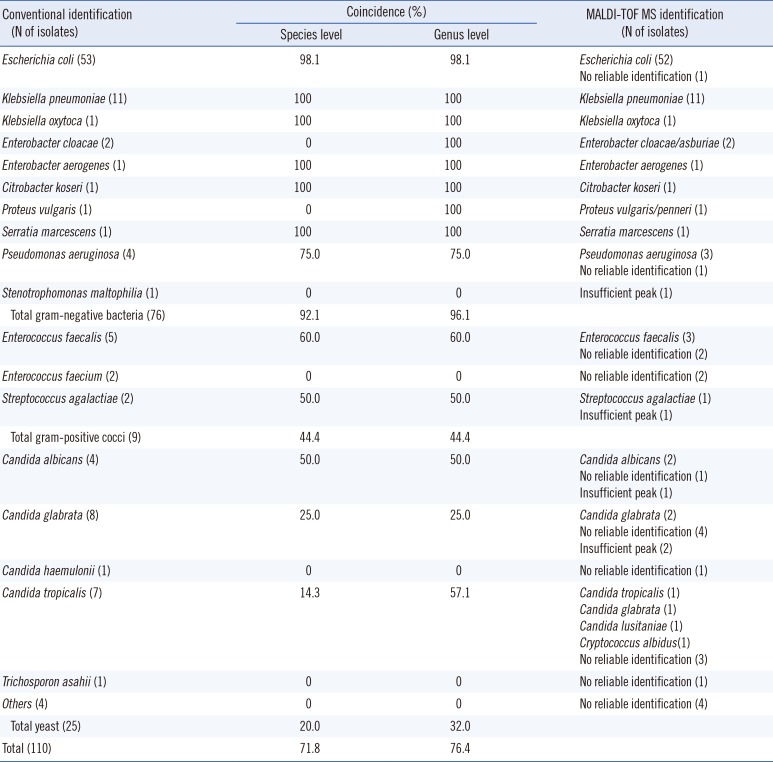
Table 3
Comparison of the results between the conventional method and MALDI-TOF in 31 specimens with colony counts of 104-105 CFU/mL




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download