Abstract
Background
ChromID Clostridium difficile agar (IDCd; bioMérieux SA, France) is a recently developed chromogenic medium for rapid and specific isolation of C. difficile. We compared the performance of IDCd with that of Clostridium difficile Selective Agar (CDSA).
Methods
A total of 530 fresh stool specimens were collected from patients with clinical signs compatible with C. difficile infection, and cultures for C. difficile were performed on IDCd and CDSA. C. difficile colonies were identified by spore staining, odor, use of an ANI identification test kit (bioMérieux SA), and multiplex PCR for tcdA, tcdB, and tpi.
Results
The concordance rate between IDCd and CDSA was 90.6% (480/530). The positivity rates on IDCd on days 1 and 2 (55.6% and 85.0%, respectively) were significantly higher than those on CDSA (19.4% and 75.6%, respectively) (P<0.001 for day 1 and P=0.02 for day 2), but the detection rates on IDCd and CDSA on day 3 were not different (89.4% vs. 82.8%, P=0.0914). On day 3, the recovery rates for non-C. difficile isolates on IDCd and CDSA were 30.2% (160/530) and 22.1% (117/530), respectively (P=0.0075). Clostridium spp. other than C. difficile were the most prevalent non-C. difficile isolates on both media.
Traditionally, Clostridium difficile culture has been used for epidemiologic studies and diagnoses of C. difficile infection (CDI). Increasing morbidity and mortality rates and the increasing frequency of recurrence and community-acquired CDI demand a reliable culture method for toxigenic C. difficile [1, 2]. The effective isolation of C. difficile from stool samples requires the use of selective media and spore selection procedures [3-8], and toxigenic culture is regarded as the standard method for detection of toxigenic C. difficile. Although bacteriologic culture is a time-consuming process requiring more than 2 days, it is more straightforward and sensitive than the cytotoxicity neutralization assay [10].
A variety of selective culture media, including cycloserine cefoxitin fructose agar (CCFA) and C. difficile selective agar (CDSA; Becton Dickinson, Sparks, MD, USA), have been used for the isolation of C. difficile [3-9]. CCFA was the first selective culture medium developed for C. difficile, but the original formulation has been modified for commercial distribution [3, 6, 8]. ChromID C. difficile agar (IDCd; bioMérieux SA, Marcy l'Etoile, France) is a recently developed chromogenic agar medium for the rapid and specific isolation of C. difficile. The aim of this study was to compare the performance of IDCd with that of CDSA for the rapid and sensitive detection of C. difficile.
A total of 530 fresh stool specimens were collected from patients who had clinical signs compatible with CDI and were hospitalized at a teaching hospital in Seoul between January and June 2012. The Institutional Review Board of Sanggye Paik Hospital approved the study protocol.
Semiquantitative culture for C. difficile was performed as described previously [10] and the extent of growth was rated as follows: grade 1, <10 colonies; grade 2, 10-50 colonies; grade 3, 51-100 colonies; and grade 4, >100 colonies. Briefly, a stool specimen (1.0 mL) was mixed with an equal volume of 70% isopropanol and incubated at room temperature for 30 min. One drop (~100 µL) was then inoculated onto prereduced IDCd or CDSA, and the plates were incubated at 37℃ under anaerobic conditions (GasPak EZ Anaerobe Pouch; Becton Dickinson) for 72 hr. We observed the plates on days 1, 2, and 3 without interrupting the anaerobic incubation. To evaluate the effect of the alcohol shock, we also inoculated 203 of the 530 stool specimens directly onto IDCd without alcohol shock pretreatment (IDCd-direct). If C. difficile was detected on either IDCd or CDSA, the specimen was defined as C. difficile positive, and any other bacteria growing on the culture media were defined as non-C. difficile isolates. C. difficile colonies were identified on the basis of typical morphological features (yellow colony with a ground-glass appearance on CDSA, black colony with an irregular margin on IDCd), spore staining, odor, and the use of an ANI identification test kit (bioMérieux SA).
Multiplex PCR for toxin A (tcdA), toxin B (tcdB), and triose phosphosphate isomerase (tpi) was performed for 180 C. difficile isolates, as described previously [11]. The PCR product for tpi was 230 bp long if the isolate was C. difficile. The PCR product for tcdA was 369 bp long if the gene was intact and 110 bp if the isolate contained the variant gene (tcdA-tcdB+). The PCR product for tcdB was 160 bp long if the gene was intact.
All statistical analyses were conducted by using SAS Version 9.2 software package (SAS Inc., Cary, NC, USA). Statistical differences in positivity rates among IDCd and CDSA groups at different days were analyzed using the generalized estimating equation (GEE) with logit link method. P values<0.05 were considered significant.
Of the 203 stool specimens cultured with and without alcohol pretreatment, 73 yielded C. difficile on at least 1 of the 2 tested media on day 3 (Table 1). The positivity rates for IDCd-direct and IDCd-alcohol on day 3 were 79.5% (58/73) and 87.7% (64/73), respectively, and were not significantly different (P=0.5630). Therefore, we further evaluated the performance of IDCd and CDSA with alcohol pretreatment.
Of the 530 alcohol-treated stool specimens used for the comparison of IDCd and CDSA, 180 were identified as C. difficile positive by day 3 (Table 2). Of the 180 C. difficile isolates, 142 (78.9%) were tcdA+tcdB+ strains, 11 (6.1%) were tcdA-tcdB+ strains, and 27 (15.0%) were tcdA-tcdB- strains; all of them were tpi positive.
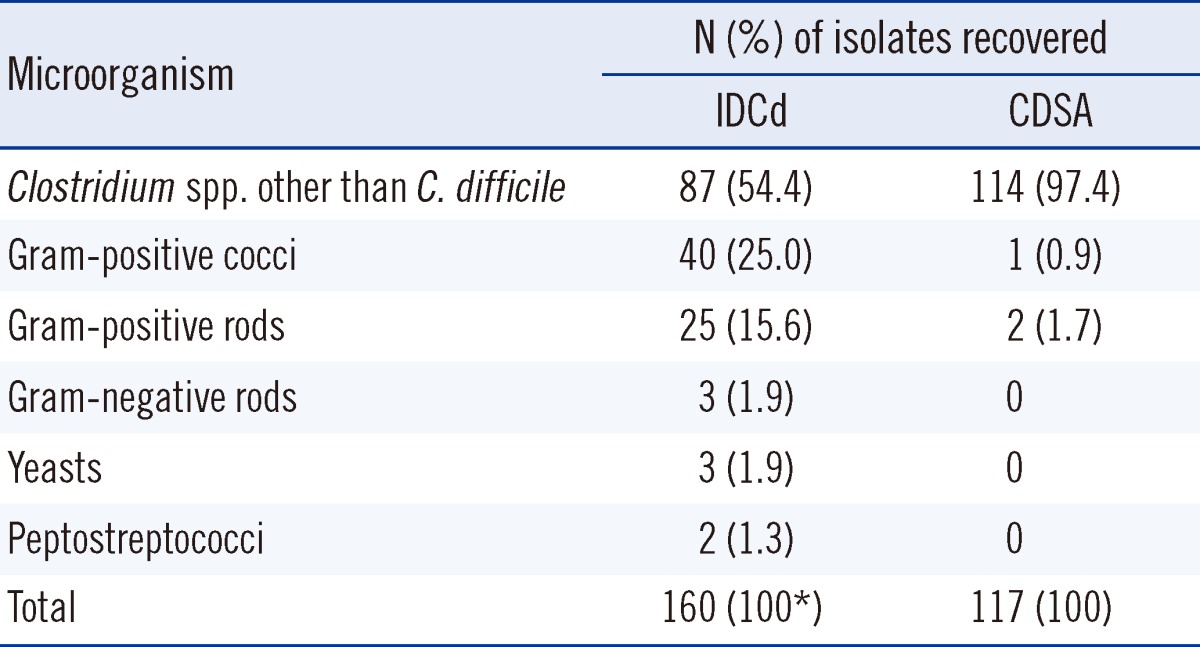
On day 3, the final C. difficile detection rates on IDCd and CDSA were 89.4% (161/180) and 82.8% (149/180), respectively, and were not significantly different (P=0.0914). However, the positivity rates on days 1 and 2 were significantly higher on IDCd than on CDSA (55.6% vs. 19.4% on day 1, 85.0% vs. 75.6% on day 2; P<0.001 for day 1, P=0.0245 for day 2). The recovery rates for non-C. difficile isolates on IDCd and CDSA on day 3 were 30.2% (160/530) and 22.1% (117/530), respectively (P=0.0075; Table 2). Clostridium spp. other than C. difficile (COd) were the most prevalent on both media (87 cases on IDCd and 114 cases on CDSA); gram-positive cocci (mostly Enterococcus spp.) were the second most prevalent type. Gram-positive bacilli, gram-negative bacilli, and yeasts were also observed on IDCd but were rare on CDSA (Table 3).
The concordance rate between IDCd and CDSA on day 3 was 90.6% (480/530). Of the 50 discordant cases, 31 were IDCd+/CDSA- and 19 were IDCd-/CDSA+. Of the 31 IDCd+/CDSA-cases, 12 were COd and 19 were not isolated on CDSA. Of the 19 IDCd-/CDSA+ cases, 10 were COd, 4 were gram-positive cocci, and 5 were not isolated on IDCd.
Following semiquantitative culture of the C. difficile positive cases (n=161 on IDCd and n=149 on CDSA), the proportions of C. difficile colonies exhibiting higher degrees of growth (grades 3 and 4) on IDCd and CDSA were 29.8% and 15.4% on day 1 (P=0.0002), 59.0% and 60.4% on day 2 (P=0.4919), and 69.6% and 72.5% on day 3 (P=0.5635), respectively (Table 4).
A variety of culture media have been described for the selective isolation of C. difficile. CCFA, which contains cefoxitin, cycloserine, and fructose, has long been the first choice among C. difficile media, but the original formulation has been slightly modified for commercial distribution [2, 6, 8]. CDSA is a bloodless agar that contains cefoxitin, cycloserine, and mannitol instead of fructose [9, 12]. Spore selection techniques are also important for effective isolation of C. difficile, and it has been reported that alcohol pretreatment of stool specimens leads to better recovery of C. difficile than heat shock techniques [7, 8].
Selective culture media for C. difficile have been evaluated by several investigators, who have shown variable recovery rates ranging from 42.6% to 99.6% depending on the type of medium, alcohol pretreatment, and/or incubation time [3-5, 8, 9, 12-15]. In our study, the positivity rates on IDCd-direct, IDCd-alcohol, and CDSA on day 3 (79.5%, 89.4%, and 82.8%, respectively) suggested reliable performance across different types of media and treatments.
A chromogenic culture medium such as IDCd enables rapid detection of C. difficile colonies. Although the final C. difficile detection rates were not significantly different between IDCd and CDSA on day 3, the positivity rate was significantly higher on IDCd than on CDSA on day 1 (55.6% vs. 19.4%, P<0.001) and on day 2 (85.0% vs. 75.6%, P=0.0245). Recent development in synthetic enzymatic substrates has allowed improved detection and identification of microorganisms in various specimens. Chromogenic culture media contain multiple substrates that allow bacteria to form colored colonies depending on their enzymatic activity. This feature facilitates the differentiation of target pathogens within polymicrobial cultures with high specificity [16, 17]. In our study, the proportion of C. difficile colonies showing a high extent of growth (grades 3 and 4) was greater on IDCd culture plates than on CDSA plates on day 1 (29.8% vs. 15.4%, P=0.0002), although the proportions were not significantly different on days 2 and 3 (Table 4). The chromogenic enzymatic substrates and suitable germinants in IDCd may allow better production of C. difficile colonies on this medium than on CDSA [4, 5, 17].
The effects of alcohol pretreatment on both media could be excluded in our study because we simultaneously applied the alcohol-pretreated stool specimens on both media. Perry et al. [15] reported that the culture positivity rate on IDCd at 24 hr was 96.3%. One plausible explanation for this difference may be a sample selection bias in the study by Perry et al. (i.e., a predominance of enzyme immunoassay [EIA]-positive samples were selected for evaluation) [15]. In our study, the culture positivity rate on CDSA after 3 days was not significantly different from that on IDCd but was much higher than the rate reported for CDSA in previous studies [9, 12]. The higher culture positivity rate in our study was probably due to alcohol pretreatment and/or the longer duration of culture [7-9]. The positivity rate was slightly higher for IDCd-alcohol than for IDCd-direct in our study, but there was no significant difference between the two results on day 3 (Table 1; 87.7% vs. 79.5%; P=0.5630).
Previous studies using IDCd or CDSA with or without alcohol pretreatment have reported that the recovery rates for non-C. difficile isolates were from 53% to 95.7%, and COd are the most prevalent non-C. difficile isolates [9, 14, 15]. In our study, the culture positivity rates for non-C. difficile isolates correlated with the duration of culture, and COd were the most prevalent non-C. difficile isolates on both media. Gram-positive cocci and gram-positive bacilli were also frequently observed on IDCd but were rare on CDSA. Nerandzic and Donskey [9] reported that Staphylococcus epidermidis and Candida spp. were also commonly recovered on CCFA and CDSA. These results indicate that IDCd and CDSA are selective but not specific for C. difficile. Thus, differentiation of C. difficile from COd is important for accurate and rapid diagnosis of C. difficile infection. On CDSA, C. difficile colonies are usually yellow with a ground-glass appearance, but colony morphological features cannot definitively differentiate C. difficile from Cod, because Cod also forms yellow ground-glass colonies on CDSA that are nearly indistinguishable from those of C. difficile [9]. Therefore, when suspicious colonies are found on CDSA, additional diagnostic procedures, such as spore staining, ANI tests, and/or PCR assays for tcdA and tcdB, must be performed [10, 11, 18]. However, on IDCd, C. difficile colonies exhibit remarkably clear and distinctive morphological features (black with irregular margins) [14, 15]. In our study, all the suspected C. difficile colonies on IDCd were tpi positive, although 15% of them were non-toxin-producing strains. The C. difficile colonies became larger and the black coloration of the colonies and the irregularity of the margins increased with increase in the duration of culture.
In conclusion, the advantage of IDCd is that although the culture positivity rates were not significantly different between IDCd and CDSA on day 3, IDCd may allow for rapid and sensitive detection of C. difficile within 2 days of cultivation (with or without alcohol pretreatment).
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2012-0008202). A part of ChromID medium was kindly provided by bioMérieux Korea.
References
1. Miller BA, Chen LF, Sexton DJ, Anderson DJ. Comparison of the burdens of hospital-onset, healthcare facility-associated Clostridium difficile infection and of healthcare-associated infection due to methicillin-resistant Staphylococcus aureus in community hospitals. Infect Control Hosp Epidemiol. 2011; 32:387–390. PMID: 21460491.
2. Shin BM, Moon SJ, Kim YS, Shin WC, Yoo HM. Characterization of cases of Clostridium difficile infection (CDI) presenting at an emergency room: molecular and clinical features differentiate community-onset hospital-associated and community-associated CDI in a tertiary care hospital. J Clin Microbiol. 2011; 49:2161–2165. PMID: 21471341.
3. Arroyo LG, Rousseau J, Willey BM, Low DE, Staempfli H, McGeer A, et al. Use of a selective enrichment broth to recover Clostridium difficile from stool swabs stored under different conditions. J Clin Microbiol. 2005; 43:5341–5343. PMID: 16208013.
4. Bliss DZ, Johnson S, Clabots CR, Savik K, Gerding DN. Comparison of cycloserine-cefoxitin-fructose agar (CCFA) and taurocholate-CCFA for recovery of Clostridium difficile during surveillance of hospitalized patients. Diagn Microbiol Infect Dis. 1997; 29:1–4. PMID: 9350408.
5. Buggy BP, Hawkins CC, Fekety R. Effect of adding sodium taurocholate to selective media on the recovery of Clostridium difficile from environmental surfaces. J Clin Microbiol. 1985; 21:636–637. PMID: 3988904.
6. George WL, Sutter VL, Citron D, Finegold SM. Selective and differential medium for isolation of Clostridium difficile. J Clin Microbiol. 1979; 9:214–219. PMID: 429542.
7. Koransky JR, Allen SD, Dowell VR Jr. Use of ethanol for selective isolation of spore forming microorganisms. Appl Environ Microbiol. 1978; 35:762–765. PMID: 348108.
8. Marler LM, Siders JA, Wolters LC, Pettigrew Y, Skitt B, Allen SD. Comparison of five cultural procedures for isolation of Clostridium difficile from stools. J Clin Microbiol. 1992; 30:514–516. PMID: 1537928.
9. Nerandzic MM, Donskey CJ. Effective and reduced-cost modified selective medium for isolation of Clostridium difficile. J Clin Microbiol. 2009; 47:397–400. PMID: 19073869.
10. Shin BM, Kuak EY, Lee EJ, Songer JG. Algorithm combining toxin immunoassay and stool culture for diagnosis of Clostridium difficile infection. J Clin Microbiol. 2009; 47:2952–2956. PMID: 19625481.
11. Lemee L, Dhalluin A, Testelin S, Mattrat MA, Maillard K, Lemeland JF, et al. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (toxin A), and tcdB (toxin B) genes for toxigenic culture of Clostridium difficile. J Clin Microbiol. 2004; 42:5710–5714. PMID: 15583303.
12. Bloedt K, Riecker M, Poppert S, Wellinghausen N. Evaluation of new selective media and a rapid fluorescence in situ hybridization assay for identification of Clostridium difficile from stool samples. J Med Microbiol. 2009; 58:874–877. PMID: 19502365.
13. Yim JS, Hwang SM, Kim M, Lim HJ, Shin S, Chung HS, et al. Evaluation of a ChromID C. difficile agar for the isolation of Clostridium difficile. Korean J Clin Microbiol. 2012; 15:88–91.
14. Hill KA, Collins J, Wilson L, Perry JD, Gould FK. Comparison of two selective media for the recovery of Clostridium difficile from environmental surfaces. J Hosp Infect. 2013; 83:164–166. PMID: 23201396.
15. Perry JD, Asir K, Halimi D, Orenga S, Dale J, Payne M, et al. Evaluation of a chromogenic culture medium for isolation of Clostridium difficile within 24 hr. J Clin Microbiol. 2010; 48:3852–3858. PMID: 20739493.
16. Perry JD, Freydière AM. The application of chromogenic media in clinical microbiology. J Appl Microbiol. 2007; 103:2046–2055. PMID: 18045388.

17. Orenga S, James AL, Manafi M, Perry JD, Pincus DH. Enzymatic substrates in microbiology. J Microbiol Methods. 2009; 79:139–155. PMID: 19679151.

18. Shin BM, Mun SJ, Yoo SJ, Kuak EY. Comparison of BD GeneOhm Cdiff and Seegene Seeplex ACE PCR assays using toxigenic Clostridium difficile culture for direct detection of tcdB from stool specimens. J Clin Microbiol. 2012; 50:3765–3767. PMID: 22952270.
Table 1
Comparison of ChromID agar (IDCd) with direct inoculation and IDCd with alcohol pretreatment for Clostridium difficile-positive cases
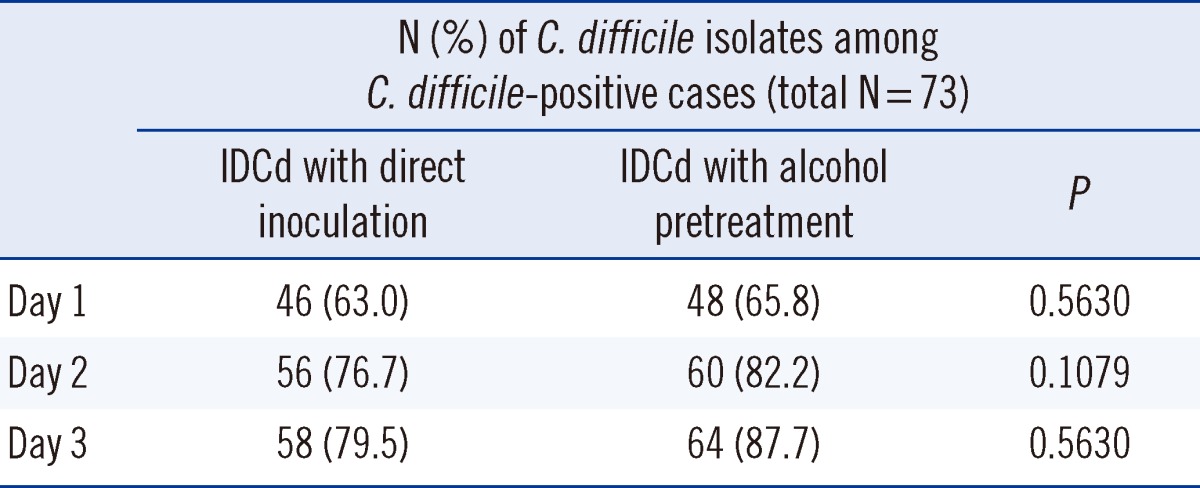
Table 2
Comparison of Clostridium difficile and non-C. difficile isolates on ChromID agar (IDCd) and C. difficile selective agar (CDSA)
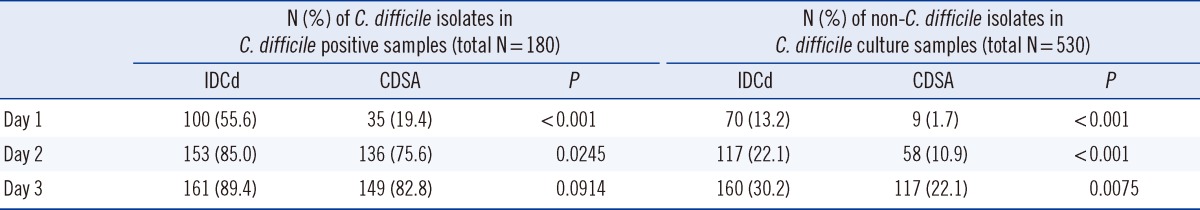
Table 3
Non-Clostridium difficile isolates from 530 stool specimens recovered on ChromID agar (IDCd) and C. difficile selective agar (CDSA) on day 3




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download