Abstract
Fanconi anemia (FA) is a rare genetic disorder affecting multiple body systems. Genetic testing, including prenatal testing, is a prerequisite for the diagnosis of many clinical conditions. However, genetic testing is complicated for FA because there are often many genes that are associated with its development, and large deletions, duplications, or sequence variations are frequently found in some of these genes. This study describes successful genetic testing for molecular diagnosis, and subsequent prenatal diagnosis, of FA in a patient and his family in Korea. We analyzed all exons and flanking regions of the FANCA, FANCC, and FANCG genes for mutation identification and subsequent prenatal diagnosis. Multiplex ligation-dependent probe amplification analysis was performed to detect large deletions or duplications in the FANCA gene. Molecular analysis revealed two mutations in the FANCA gene: a frameshift mutation c.2546delC and a novel splice-site mutation c.3627-1G>A. The FANCA mutations were separately inherited from each parent, c.2546delC was derived from the father, whereas c.3627-1G>A originated from the mother. The amniotic fluid cells were c.3627-1G>A heterozygotes, suggesting that the fetus was unaffected. This is the first report of genetic testing that was successfully applied to molecular diagnosis of a patient and subsequent prenatal diagnosis of FA in a family in Korea.
Fanconi anemia (FA) is a rare genetic disorder characterized by variable congenital anomalies, progressive bone marrow failure, and high predisposition to acute leukemia and other malignancies [1-4]. FA shows severe genetic heterogeneity, although the proteins encoded by FA-related genes are considered to work together in a common pathway that regulates cellular resistance to DNA cross-linking agents [2]. At least 15 genes have been identified that are responsible for FA complementation groups: FANCA, FANCB, FANCC, BRCA2 (FANCD1), FANCD2, FANCE, FANCF, FANCG (XRCC9), FANCI, BRIP1 (FANCJ or BACH1), FANCL, FANCM, PALB2 (FANCN), RAD51C (FANCO), and SLX4 (FANCP)[5, 6]. Abnormalities of FA genes are inherited in an autosomal recessive manner, except for FANCB mutations, which are inherited in an X-linked manner. Molecular diagnosis of FA is quite complicated, not only because at least 15 genes are associated with its development, but also the mutation spectra of most FA-associated genes are very diverse and some of these genes frequently contain large deletions or duplications [7-9].
Mutational information is a prerequisite for genetic counseling of family members, screening of potential bone marrow transplantation donors who are phenotypically and hematologically normal, and prediction of clinical prognosis on the basis of genotype-phenotype correlations. We describe the successful application of genetic testing to the molecular diagnosis of FA, and subsequently to prenatal diagnosis of FA, in a patient and his family in Korea.
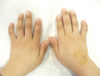
A 4-yr-old male presented at the hospital with a chief medical complaint of recurrent abdominal pain and hematochezia. The patient was the first child of unrelated healthy parents born after 41weeks of pregnancy. The patient had a history of recurrent pneumonia, epistaxis, easy bruising, urinary urgency, and perineal area pain. Physical examination of the patient revealed short stature, clinodactyly with brachymesophalangia on bilateral 5th fingers, multiple café-au-lait spots on the right knee, left thigh, pelvis, and right buttock (Fig. 1). He had no eyeball abnormalities or ear problems. Urological examination was unremarkable. His initial complete blood cell count results were as follows: white blood cell, 4.0×109/L; hemoglobin, 12.5 g/dL; platelets, 78×109/L. Repeat complete blood cell counts indicated persistent thrombocytopenia. No bone marrow examination was included in the initial study. No family members (both parents and a younger sister) had experienced symptoms and manifestations that were similar to those of the patient. At the time of the initial examination, the patient's mother was pregnant with her third child.
A standard chromosomal breakage test with diepoxybutane (DEB) and mitomycin C (MMC) was performed, revealing chromosomal hypersensitivity to clastogenic agents. The mean number of breaks per metaphase, the ratio of the mean number of breaks per metaphase in patient/control, and the number of chromosome breaks per aberrant mitosis were higher than the normal range for non-FA cells [10] (Fig. 2). The patient's clinical and cytogenetic findings were compatible with FA.
Up to 85% of FA cases are attributable to the 3 most common FA genes: FANCA, FANCC, and FANCG. The patient underwent genetic testing for these genes to identify causative mutations and to prenatally diagnose the fetus at 16+3 gestational weeks. A Puregene DNA isolation kit (Gentra Systems Inc., Minneapolis, MN, USA) was used to extract genomic DNA from peripheral blood leukocytes of the proband, his parents, and his sister, as well as from cultured amniotic fluid cells, according to the manufacturer's protocol. PCR was performed to amplify the entire coding and flanking regions of all 72 exons of the FANCA, FANCC, and FANCG genes. The primers were designed using Primer3 PLUS (http://www.bioinformatics.nl/cgi-din/primer3plus/primer3plus.cgi) and the reference sequences of FANCA (NC_000016.9, NM_000135.2), FANCC (NC_000009.11, NM_000136.2), and FANCG (NC_000009.11, NM_004629.1). Base pair number +1 was assigned to the A of the ATG translation initiation start site for reference. Amplified products were sequenced bi-directionally in an ABI PRISM 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) using BigDye Terminator v3.1 Cycle sequencing kits (Applied Biosystems), then analyzed using Seqscape software (Applied Biosystems).
Reverse transcription-PCR (RT-PCR) was performed to determine the mutational effects of a potential splice-site mutation. The RT-PCR fragments were analyzed by 2% agarose gel electrophoresis and capillary electrophoresis using the Labchip GX Caliper (Caliper Life Sciences, Hopkinton, MA, USA). All RNA products were directly sequenced using the ABI PRISM 3730 Genetic Analyzer. We determined the allele frequencies in 95 control subjects and performed in silico prediction to estimate the significance of novel missense variants using 3 software programs: PolyPhen (http://genetics.bwh.harvard.edu/pph/), Align-GVGD (http://agvgd.iarc.fr), and SIFT (http://sift.jcvi.org).
We also performed multiplex ligation-dependent probe amplification (MLPA) to detect large deletions or duplications within the FANCA gene using a SALSA P031-A2/P032 kit (MRC-Holland BV, Amsterdam, The Netherlands) [11]. PCR products were analyzed in an ABI PRISM 3130 Genetic analyzer (Applied Biosystems) and the data were analyzed using GeneMarkerver. 1.51 (Softgenetics, State College, PA, USA). Peak heights were normalized, and a deletion or duplication was suspected when the normalized peak ratio was less than 0.75 or greater than 1.30.
Targeted mutational analysis was performed for the patient's family members to determine whether they also had the mutations identified in the proband. To exclude the possibility of maternal cell contamination into fetal amniotic fluid cells, genotyping of samples from the mother and the amniotic fluid cells was performed for 10 short tandem repeat (STR) loci using the AmpFlSTR Profiler Plus PCR amplification kit (Applied Biosystems), which co-amplifies the loci D3S1358, vWA, FGA, D8S1179, D21S11, D18S51, D5S818, D13S317, D7S820, and amelogenin.
The molecular study revealed compound heterozygous mutations in the FANCA gene of the proband. One mutation was a previously reported frameshift mutation, c.2546delC (p.Ser849 Phefs*40), whereas the other was a novel splice-site mutation, c.3627-1G>A, in intron 36 (Fig. 3). A subsequent RNA study identified that this G-to-A substitution at the splicing acceptor site in intron 36 results in aberrant splicing, which leads to skipping of the first 16-bp of exon 37, and finally to a shift in the reading frame (p.Asp1209Glufs*33) (Fig. 4). Additionally, 2 sequence variations in the FANCA gene, c.3031C>T (p.Arg1011Cys) and c.3472A>G (p.Lys1158Glu), were identified. These were predicted to be benign using in silico approaches. Ten homozygote polymorphisms in the FANCA gene, c.710-12A>G, c.796A>G, c.1143G>T, c.1226-20A>G, c.1501G>A, c.1826+15T>C, c.2151+8T>C, c.2426G>A, c.3935-16C>T, and c.154G>A were also identified. No large deletions or duplications were identified within the FANCA gene. No other specific mutations were found in the FANCC or FANCG genes. Only 3 homozygote polymorphisms in the FANCG gene were found: c.-490G>T, c. -453_-452insT, and c.-392A>G.
The father of the patient had a heterozygous c.2546delC mutation, whereas the patient's mother carried a heterozygous c.3627-1G>A mutation (Fig. 3). No mutation was identified in the patient's younger sister. Only the heterozygous c.3627-1G>A mutation was identified in the DNA extracted from amniotic fluid cells. STR analysis revealed the presence of the Y chromosome in amniotic fluid cells and distinct genotypes among maternal blood cells and amniotic fluid cells in loci D3S1358, D5S818, vWA, and FGA (Fig. 5).
This is the first FA case to be genetically confirmed in Korea. Additionally, this is the first report of genetic testing that was successfully applied for subsequent prenatal diagnosis of FA in a patient's family in Korea. A few previous cases of FA are reported to have been diagnosed using only the chromosome breakage test [12].
The characteristic cellular phenotype of FA includes chromosomal instability and hypersensitivity to DNA cross-linking agents [13, 14]. The diagnosis of FA is usually based on clinical suspicion and chromosomal hypersensitivity to DNA cross-linking agents [15]. However, the chromosome breakage test can sometimes generate false-negative results, similar to that observed in cases of somatic mosaicism [16]. Furthermore, prenatal chromosome breakage studies may be inconclusive and inaccurate because of the variable growth status of amniotic cells and the poor yield of cells in metaphase [17]. Although genetic testing is the method of choice for prenatal diagnosis, genetic testing is not widely used in the diagnosis and management of FA. This is partly due to the severe genetic heterogeneity of FA and the technical difficulties associated with identifying diverse mutations in FA genes. Therefore, technical advances in DNA analysis, such as next-generation sequencing technology, can facilitate the performance of simple and easy genetic tests for FA.
Figures and Tables
Fig. 2
Cytogenetic findings of a chromosome breakage test with (A) DEB (diepoxybutane) and (B) MMC (mitomycin C) in a peripheral blood lymphocyte culture. Increased mean number of breaks per metaphase (DEB: 5.5, MMC: 11.2, FA cutoff >2.0), ratio of mean number of breaks per metaphase in patient/control (DEB: 275, MMC: 112, FA cutoff >10), and number of chromosome breaks per aberrant mitosis (DEB: 6.05, MMC: 11.2, FA cutoff >5.5) are recorded.

Fig. 3
Sequencing results of the FANCA gene show compound heterozygous mutations of c.2546delC (denoted by an asterisk; A) and c.3627-1G>A (denoted by an asterisk; B) in the proband. The c.2546delC mutation was inherited paternally, whereas the c.3627-1G>A mutation was transmitted maternally. The fetus was a heterozygous carrier of the c.3627-1G>A mutation.

Fig. 4
Results of agarose gel electrophoresis of RT-PCR products (A), capillary electrophoresis of RT-PCR products (B), and sequence analysis (C) for the novel potential splice-site mutation c.3627-1G>A. RT-PCR was conducted using following primers: F-5'-TTGACCTCTGCTCTGGTGTG-3', R-5'-AACCAATAGCTCCTCTCTCTCG-3'. In addition to the normal transcript (276-bp-sized band), an abnormal transcript (260-bp-sized band) was discernible on capillary electrophoresis. Sequence analysis demonstrated that this abnormal transcript resulted from skipping of the first 16 bp (yellow) of exon 37 rather than all of exon 37 (blue).
Abbreviation: RT-PCR, reverse transcription PCR.
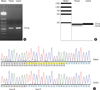
Fig. 5
Results of STR marker analysis that was performed to determine maternal cell contamination into amniotic fluid. For the FGA marker, maternal blood has allele 20.3 and allele 24.3, while the amniotic fluid has only allele 20.3 (A). For the D3S1358 marker, amniotic fluid has allele 15.1, which is not shared with maternal blood, as well as allele 16.2 (B).
Abbreviation: STR, short tandem repeat.

References
1. Alter BP. Diagnosis, genetics, and management of inherited bone marrow failure syndromes. Hematology Am Soc Hematol Educ Program. 2007. 29–39.

2. Taniguchi T, D'Andrea AD. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. 2006. 107:4223–4233.

3. Kutler DI, Singh B, Satagopan J, Batish SD, Berwick M, Giampietro PF, et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR). Blood. 2003. 101:1249–1256.

4. Rosenberg PS, Greene MH, Alter BP. Cancer incidence in persons with Fanconi anemia. Blood. 2003. 101:822–826.

5. Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010. 42:406–409.

6. Stoepker C, Hain K, Schuster B, Hilhorst-Hofstee Y, Rooimans MA, Steltenpool J, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011. 43:138–141.

7. Wijker M, Morgan NV, Herterich S, van Berkel CG, Tipping AJ, Gross HJ, et al. Heterogeneous spectrum of mutations in the Fanconi anaemia group A gene. Eur J Hum Genet. 1999. 7:52–59.

8. Bouchlaka C, Abdelhak S, Amouri A, Ben Abid H, Hadiji S, Frikha M, et al. Fanconi anemia in Tunisia: high prevalence of group A and identification of new FANCA mutations. J Hum Genet. 2003. 48:352–361.
9. Callén E, Tischkowitz MD, Creus A, Marcos R, Bueren JA, Casado JA, et al. Quantitative PCR analysis reveals a high incidence of large intragenic deletions in the FANCA gene in Spanish Fanconi anemia patients. Cytogenet Genome Res. 2004. 104:341–345.

10. Soulier J, Leblanc T, Larghero J, Dastot H, Shimamura A, Guardiola P, et al. Detection of somatic mosaicism and classification of Fanconi anemia patients by analysis of the FA/BRCA pathway. Blood. 2005. 105:1329–1336.

11. Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002. 30:e57.

12. Kook H, Cho D, Cho SH, Hong WP, Kim CJ, Park JY, et al. Fanconi anemia screening by diepoxybutane and mitomicin C tests in Korean children with bone marrow failure syndromes. J Korean Med Sci. 1998. 13:623–628.

13. Yamashita T, Nakahata T. Current knowledge on the pathophysiology of Fanconi anemia: from genes to phenotypes. Int J Hematol. 2001. 74:33–41.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download