Abstract
Background
Acinetobacter baumannii resistance islands (AbaRs) have been recently recognized as mobile genetic elements that harbor multiple resistance determinants and are associated with multidrug resistance (MDR). In the present study, we aimed to determine the AbaRs conferring multiple antimicrobial resistance and their clonal relatedness to MDR A. baumannii clinical isolates obtained from a university hospital in Daejeon, Korea.
Methods
This study included 29 MDR A. baumannii strains isolated in Daejeon, Korea. The minimal inhibitory concentrations (MICs) were determined by Etest. A. baumannii isolates were characterized using the 2 multiplex PCR assays and multilocus sequence typing (MLST) scheme. To detect and characterize AbaRs, PCR and PCR mapping experiments were performed.
Results
Twenty-seven of the 29 isolates belonged to the European (EU) clone II lineage and contained 5 sequence types (STs) (75, 92, 137, 138, and 357). In this study, ST357 was confirmed for the first time in Korea. Only 2 of the 29 isolates belonged to the EU clone I lineage, and were confirmed as ST109. These 2 isolates harbored the 22-kb AbaR7 aacC1-orfP-orfQ-aadA1 gene cassette array. In contrast, AbaR was not found in EU clone II isolates.
Acinetobacter baumannii is an aerobic, glucose-nonfermenting, Gram-negative bacterium that has recently emerged as a serious opportunistic and nosocomial pathogen [1]. A few lineages of multidrug-resistant (MDR) A. baumannii strains, which are resistant to all or most clinically relevant antimicrobial agents, have been reported worldwide and have caused multiple hospital outbreaks. In particular, 2 of these lineages, European (EU) clones I and II, have become widespread worldwide [2].
It has been suggested that MDR A. baumannii strains acquire their antimicrobial resistant genes via resistance islands, integrons, and transposons that carry 1 or more antimicrobial resistance gene cassettes [3]. However, despite extensive research, not much is known about the role of these genetic mobile elements in the evolution of MDR A. baumannii. Recent studies have revealed that some A. baumannii strains harbor multiple antimicrobial resistance regions that are integrated into the comM gene, which encodes an ATPase domain. The first example of these regions, which was found in strain A. baumannii AYE, was designated A. baumannii resistance island (AbaR)1 and harbors 45 genes putatively associated with resistance to antimicrobial agents or biocides. These antimicrobial resistance genes confer resistance to aminoglycosides (kanamycin, gentamicin, and neomycin), aminocyclitols (spectinomycin and streptomycin), tetracycline, and chloramphenicol [4].
Following detection of the 86.2-kb AbaR1, 9 additional AbaRs have been fully characterized. All but 1 AbaR (AbaR1, AbaR3, AbaR5, AbaR6, AbaR7, AbaR8, AbaR9, and AbaR10) are found in EU clone 1 strains and have a complex structure, a 16.3-kb backbone transposon (Tn6019) disrupted by a cadmium and zinc resistance gene, and a second transposon (Tn6018) interrupted by a variable assortment of antimicrobial resistance genes. In addition, the blaoxa-23 gene-carrying AbaR4, which is integrated at a chromosomal site other than the comM gene, has been identified in some EU clone I and EU clone II strains [2]. AbaR2 in EU clone II strains consists of a largely truncated AbaR that contains only the right-hand part of an AbaR island and a transposon related to Tn6021. There have been much fewer reports of AbaRs in EU clone II strains.
Although AbaRs have been recently recognized as mobile genetic elements that harbor multiple resistance determinants and are associated with MDR in A. baumannii, there is a relative paucity of data on the number and types of AbaRs in MDR A. baumannii strains isolated from Korea. In the present study, we aimed to determine the AbaRs associated with resistance to multiple antimicrobials and their clonal relatedness to the MDR A. baumannii clinical isolates obtained from a university hospital in Daejeon, Korea.
Twenty-two MDR A. baumannii isolates were collected and characterized [5]. Table 1 shows the minimal inhibitory concentrations (MICs) and antimicrobial resistance determinants of the isolates characterized in this study. These isolates were collected from different patients at a single university hospital in Daejeon, Korea during the period between 2007 and 2011. Seven additional strains that were susceptible to carbapenem but resistant to many other antimicrobial agents were also included in this study. Piperacillin, piperacillin/tazobactam, cefepime, ceftazidime, meropenem, and ticarcillin susceptibility testing was performed using the Vitek 2 system (bioMérieux, Marcy l'Eltoile, France). The MICs of imipenem, amikacin, gentamicin, and ciprofloxacin were determined using the Etest (AB Biodisk, Solna, Sweden). Interpretation was performed according to the criteria approved by the CLSI guidelines [6]. Escherichia coli ATCC 25922 was used as a reference strain. Clinical isolates of A. baumannii were identified by rpoB gene analysis and by the presence of the blaoxa-51-like gene [7].
The MDR phenotype was defined as resistance to representative antimicrobial agents of at least 3 different classes of drugs: aminoglycosides (gentamicin, amikacin), antipseudomonal penicillins (ticarcillin, piperacillin, piperacillin/tazobactam), carbapenems (imipenem, meropenem), antipseudomonal cephalosporins (ceftazidime, cefepime), and fluoroquinolones (ciprofloxacin) [8].
Whole-cell (genomic) DNA was obtained from each target strain using a genomic DNA purification kit (SolGent, Daejeon, Korea) according to the manufacturer's instructions. PCR was performed using 50 ng of genomic DNA, 2.5 µL of 10× Taq buffer, 0.5 µL of 10 mM dNTP mix, 20 pmol of each primer, and 0.7 U of Taq DNA polymerase (SolGent) in a total volume of 25 µL. Each target gene was amplified in a GeneAmp PCR System 9600 thermal cycler (Perkin-Elmer Cetus Corp., Norwalk, CT, USA). Thermal cycling conditions consisted of an initial denaturation cycle at 95℃ for 5 min, followed by 30 cycles of 95℃ for 30 sec, 52℃ for 40 sec, and 72℃ for 30 sec, with a final extension at 72℃ for 5 min. The annealing temperature was 52℃, unless otherwise specified. The amplified products were separated via electrophoresis on 1.5% (w/v) agarose gels containing ethidium bromide, and visualized using a BioDoc-14TM Imaging system (UVP, Cambridge, UK). For sequencing, PCR products were purified with a PCR purification kit (SolGent) according to the manufacturer's protocols.
The 2 multiplex PCR assays were performed as previously described [9] to identify members of the EU clone I and EU clone II lineages. Epidemiological typing of the isolates was performed by repetitive extragenic palindromic sequence (REP) PCR [10]. The Oxford multilocus sequence typing (MLST) scheme [11], which uses 7 housekeeping genes (gltA, gyrB, gdhB, recA, cpn60, gpi, and rpoD24) was used to determine the sequence types (STs). A ST number was assigned by comparing the allele sequences to those on the MLST site (http://pubmlst.org/abaumannii/). In addition, class 1 integrons were detected and sequenced using PCR conditions and a primer described previously [5].
The genes associated with the AbaR islands were detected by PCR using published primers (Table 2). The amplified regions are shown in Fig. 1 [4]. To investigate the structure of the AbaR backbone, PCR mapping experiments were performed as described previously [4]. The amplicons were purified and sequenced using a BigDye Terminator Cycle Sequencing Kit (PE Applied Biosystems, Foster City, CA, USA) and an ABI PRISM 3730XL DNA analyzer (PE Applied Biosystems). DNA fragments (up to 1 kb in size) were sequenced using the overlapping PCR technique. The various DNA sequences were confirmed using the Basic Local Alignment Search Tool (BLAST) program (http://blast.ncbi.nlm.gov).
The 29 MDR A. baumannii strains were tested to determine their association with the EU clone lineages using multiplex PCRs. Twenty-seven isolates belonged to the EU clone II lineage and carried allele 66 of the intrinsic blaOXA-51-like genes, which corresponds to their assignment to the EU clone II lineage. MLST analysis of the EU clone II isolates revealed 5 STs (75, 92, 137, 138, and 357) (Table 3). In particular, ST357 (1-12-3-2-2-145-3) was confirmed for the first time in Korea. The strains identified as ST357 were susceptible to imipenem although they were MDR strains (Table 1). Among the 29 MDR A. baumannii strains, only 2 isolates belonged to the EU clone I lineage and contained allele 69 of the intrinsic blaOXA-51-like genes, which corresponds to their assignment to the EU clone I lineage. The 2 isolates were confirmed as ST109 (10-12-4-11-4-9-5) by MLST analysis.
Most of the MDR A. baumannii isolates (93.1%) contained 2.5-kb class 1 integrons, and the gene cassette arrays were divided into 2 types by nucleotide sequencing. The gene cassette array aacA4-catB8-aadA1 was detected in only the EU clone II isolates, whereas all EU clone I isolates only carried the gene cassette array aacC1-orfP-orfQ-aadA1.
To determine the clonality, REP-PCR was performed; the 29 MDR A. baumannii isolates displayed only 2 REP-PCR patterns, designated type I and type II (Fig. 2). The 2 EU clone I isolates exhibited the type I pattern, while the 27 EU clone II isolates exhibited the type II pattern.
The comM gene was not detected in any of our isolates, but was amplified from the Acinetobacter calcoaceticus control strain.
Segments J1 and J2, which form the boundaries of the AbaR backbone transposon Tn6019, were amplified from all 29 isolates (both EU clone I and EU clone II), whereas Tn6018 and an interrupted uspA gene were only present in the EU clone I isolates. These results indicated that an AbaR was only present in the EU clone I isolates. However, it is important to note that J4 (the junction of Tn6018-R with Tn6019) was amplified, but J3 (the junction of Tn6018-L with Tn6019) was not amplified in the EU clone I isolates. In addition, only J6 of the segments J5 and J6 (the junctions of multiple-antibiotic resistance region [MARR] with Tn6018) was amplified in these isolates.
To characterize the AbaRs contained in the EU clone I isolates, we mapped the continuous regions of J1, J2, J4, and J6 by overlapping PCRs and sequencing. The 22-kb AbaR7 (GenBank accession no. GQ406246) was identified in the genome of the MDR A. baumannii isolates that belonged to the EU clone I lineage and were confirmed as ST109 (Fig. 3). The MARR was located between J1 and J6, and harbored a class 1 integron-containing aacC1-orfP-orfQ-aadA1 gene cassette array. The aacC1 gene conferred resistance to gentamicin, and the aadA1 gene conferred resistance to streptomycin and spectinomycin. The aphA1 gene, the kanamycin and neomycin resistance gene, was flanked by directly oriented copies of IS26.
Most EU clone I and EU clone II isolates are resistant to many antimicrobial agents that are currently used for treatment, and are important opportunistic pathogens associated with life-threatening nosocomial infections and hospital outbreaks. In this study, 27 of 29 MDR A. baumannii strains belonged to the EU clone II lineage and had identical REP-PCR patterns, indicating the clonal relationship and horizontal spread of EU clone II isolates in Daejeon, Korea. These MDR A. baumannii strains have been reported worldwide, including in Korea [9, 12].
In particular, 2 EU clone I isolates (ST109) identified in the present study were isolated in 2007 and 2012. This result suggests that EU clone I isolates (ST109) have existed in Korea for many years even though there have not been any previous reports on the isolation on EU clone I strains in Korea [12]. This is the first report of ST109 A. baumannii strains in Korea. ST109 isolates have been also recovered in Algeria, Argentina, Bulgaria, the UK, and the Netherlands, indicating their global dissemination [13]. In addition, we found a relationship between REP-PCR patterns and isolates that belong to the EU clone I lineage, which contain the blaOXA-69 gene, or the EU clone II lineage, which contain the blaOXA-66 gene.
Two types of class 1 integrons were detected in the MDR A. baumannii isolates in our study. It appeared that the integron with the aacA4-catB8-aadA1 gene cassette array was confirmed in only the EU clone II isolates; however, the integron with the aacC1-orfP-orfQ-aadA1 gene cassette array was detected in only the EU clone I isolates (ST109). In addition, the integron with the aacA4-catB8-aadA1 gene cassette array has been reported in not only A. baumannii, but also in many other bacteria, including Klebsiella pneumoniae, Citrobacter freundii, and Salmonella enterica [14]. In particular, the gene cassette array aacC1-orfP-orfQ-aadA1 is known to be located in AbaR regions and is typical of EU clone I isolates [4].
The structure of AbaRs was analyzed by a strategy based on the sequence and structural homology of the AbaRs. Although an interrupted comM gene was detected in all EU clone II isolates, the uspA gene was uninterrupted and Tn6018 was not found. Our results suggest that transposon insertion in the EU clone II strains was not closely connected to the Tn6018-containing AbaRs. However, the comM and uspA genes were interrupted and Tn6018 was detected in the EU clone I isolates (ST109), indicating that AbaRs should be present in these strains.
The AbaRs detected in present study were all confirmed as AbaR7 carrying a class 1 integron with the aacC1-orfP-orfQ-aadA1 gene cassette array. AbaR7 was recovered in an EU clone I strain isolated in Australia in 2005 [4], but has not yet been detected in A. baumannii isolates from Korea. In the EU clone I isolates (ST109), we found an AbaR7 that lacked a large internal region, including the left portion of Tn6018 and part of the Tn6019 backbone when compared to AbaR3, the original genomic structure of AbaR detected in the EU clone I lineage thus far (Fig. 3). In contrast to our results, an ST109 isolate was reportedly recovered from the Netherlands in 1984, which harbored AbaR11. These results indicate that the identical type of AbaR may be contained in varied strains of A. baumannii. Diverse AbaRs (8 types) were also detected in ST1 A. baumannii strains isolated from hospitals in the Czech Republic, Italy, and the UK [2].
AbaRs (except AbaR2) have been previously identified in EU clone I isolates, and we also detected AbaR7 only in EU clone 1 isolates. Although AbaR2 and AbaR4 were recently found in EU clone II isolates [15-17], our examination did not show AbaRs in any EU clone II isolates. Consequently, various AbaRs in the EU clone I and EU clone II lineages seem to play a substantial role in antimicrobial resistance in MDR A. baumannii isolates.
In present study, it was found that EU clone I isolates contained AbaR7 with a class 1 integron carrying antimicrobial resistance genes. However, further investigation is required to recover various AbaRs in MDR A. baumannii strains isolated from Korea. The EU clone I (ST109) isolates in this study were resistant to multiple antimicrobial agents, although they were susceptible to carbapenem. This result suggests that EU clone I isolates have been rarely recovered in Korea because previous studies focused mainly on carbapenem resistant or non-susceptible A. baumannii isolates. This finding emphasizes the idea that the antimicrobial resistance mechanisms enabling the development of multidrug resistance should be investigated not only in carbapenem-resistant MDR A. baumannii isolates, but also carbapenem-susceptible isolates.
Figures and Tables
Fig. 1
Schematic representation of A. baumannii resistance (AbaR) region showing the boundaries between interrupted genes. J1, J2, J3, J4, J5, and J6 represent the amplification regions used in the diagnostic PCRs.
Abbreviations: AbaR, A. baumannii resistance; MARR, multiple-antibiotic resistance region.

Fig. 2
Repetitive element sequence-based (REP)-PCR patterns of multidrug-resistant A. baumannii strains. Lane M, 1-kb DNA size marker.
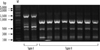
Fig. 3
Schematic representation of AbaR3 (A) and AbaR7 (B) isolated from multidrug-resistant A. baumannii strains that belonged to the EU clone I lineage. The dotted line in AbaR3 represents the deleted portion in AbaR7. The horizontal arrows indicate the orientation of gene translation.
Abbreviations: AbaR, A. baumannii resistance; MARR, multiple-antibiotic resistance region.

Table 1
The MICs of antimicrobial agents and characteristics of MDR A. baumannii isolates

*Indicates sense mutations at the 83rd residue (resulting in a serine to leucine change) in gyrA, and at the 80th residue (resulting in a serine to leucine or tryptophan change) in parC.
Abbreviations: MIC, minimum inhibitory concentration; MDR, multidrug resistance; ST, sequence type; AMK, amikacin; GEN, gentamicin; IPM, imipenem; CIP, ciprofloxacin; AMEs, aminoglycoside-modifying enzymes.
Acknowledgement
This study was financially supported by research fund of Chungnam National University 2011.
References
1. Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008. 21:538–582.
2. Krizova L, Dijkshoorn L, Nemec A. Diversity and evolution of AbaR genomic resistance islands in Acinetobacter baumannii strains of European clone I. Antimicrob Agents Chemother. 2011. 55:3201–3206.
3. Petersen A, Guardabassi L, Dalsgaard A, Olsen JE. Class I integrons containing a dhfrI trimethoprim resistance gene cassette in aquatic Acinetobacter spp. FEMS Microbiol Lett. 2000. 182:73–76.
4. Post V, White PA, Hall RM. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2010. 65:1162–1170.
5. Sung JY, Kwon KC, Cho HH, Koo SH. Antimicrobial resistance determinants in imipenem-nonsusceptible Acinetobacter calcoaceticus-baumannii complex isolated in Daejeon, Korea. Korean J Lab Med. 2011. 31:265–270.
6. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Twentieth informational supplement, M100-S20. 2010. Wayne, PA: Clinical and Laboratory Standards Institute.
7. Ko KS, Suh JY, Kwon KT, Jung SI, Park KH, Kang CI, et al. High rates of resistance to colistin and polymyxin B in subgroups of Acinetobacter baumannii isolates from Korea. J Antimicrob Chemother. 2007. 60:1163–1167.
8. Lee K, Yong D, Jeong SH, Chong Y. Multidrug-resistant Acinetobacter spp.: increasingly problematic nosocomial pathogens. Yonsei Med J. 2011. 52:879–891.
9. Turton JF, Gabriel SN, Valderrey C, Kaufmann ME, Pitt TL. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin Microbiol Infect. 2007. 13:807–815.
10. Bou G, Cervero G, Dominguez MA, Quereda C, Martinez-Beltran J. PCR-based DNA fingerprinting (REP-PCR, AP-PCR) and pulsed field gel electrophoresis characterization of a nosocomial outbreak caused by imipenem- and meropenem-resistant Acinetobacter baumannii. Clin Microbiol Infect. 2000. 6:635–643.
11. Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodríguez-Valera F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol. 2005. 43:4382–4390.
12. Higgins PG, Dammhayn C, Hackel M, Seifert H. Global spread of carbapenem-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2010. 65:233–238.
13. Hamidian M, Hall RM. AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J Antimicrob Chemother. 2011. 66:2484–2491.
14. Turton JF, Kaufmann ME, Glover J, Coelho JM, Warner M, Pike R, et al. Detection and typing of integrons in epidemic strains of Acinetobacter baumannii found in the United Kingdom. J Clin Microbiol. 2005. 43:3074–3082.
15. Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, et al. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol. 2008. 190:8053–8064.
16. Shaikh F, Spence RP, Levi K, Ou HY, Deng Z, Towner KJ, et al. ATPase genes of diverse multidrug-resistant Acinetobacter baumannii isolates frequently harbor integrated DNA. J Antimicrob Chemother. 2009. 63:260–264.
17. Mugnier PD, Poirel L, Naas T, Nordmann P. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg Infect Dis. 2010. 16:35–40.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download