Abstract
Background
Nontyphoid Salmonella (NTS) is a leading cause of human food-borne enteritiS. It has been known that integron, a naturally occurring gene capture and expression element, plays an important role in the development and dissemination of multidrug-resistance. In this study, we investigated the prevalences and molecular characteristics of integrons in NTS clinical strains.
Materials and Methods
Between 1995-96 and 2000-03, a total 261 NTS clinical strains comprising 39 serotypes were collected from clinical specimens. All strains were serotyped, and the MICs of ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, tetracycline, and trimethoprim were determined by agar dilution method. Integrons were detected by PCR amplification of integrase genes, and gene cassettes were determined by PCR and sequencing. Conjugation experiments were performed using E. coli J53 as a recipient. The clonal relationship was analyzed by pulsed-field gel electrophoresis (PFGE).
Results
Of the 261 strains tested, class 1 integrons were present in 21 strains (8.0%). Class 2 and class 3 integrons were not found. The integron-positive rate was higher in S. Typhimurium (24.2% [8/33]) than in S. Enteritidis (2.0% [3/153]). Overall rates of antimicrobial resistance were higher in integron-positive strains. dhfr12-orfF-aadA2 gene cassette was detected in 5 strains, aadA2 in 4 strains, dhfr17-orfF-aadA5 in 2 strains, and addA1 in 1 strain. Ten integron-positive transconjugants were successfully selected. Among 8 integron-positive strains of S. Typhimurium, 7 had similar PFGE patterns.
Figures and Tables
Figure 1
Polymerase chain reaction fragments from the cassette regions of class 1 integrons. M, Molecular weight Marker (1 kb ladder); S. Enteritidis (Lanes 1-3); S. Typhimurium (Lanes 4-11); S. Heidelberg (Lanes 12, 13); S. Montevideo (Lanes 14, 15); S. Dubulin (Lane 16); S. Sinstorf (Lane 17); S. Cuckmere (Lane 18); S. salamae (Lane 19); S. Othmarschen (Lane 20); S. Haifa (Lane 21).
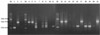
Figure 2
Pulsed-field gel electrophoresis patterns of chromosomal DNA restriction fragments resolved in 1.0% Seakem Gold agarose in 0.5×TBE buffer for Salmonella DNA digestion with XbaI. M, ladder size marker; S. Enteritidis (Lanes 1-3); S. Typhimurium (Lanes 4-11); S. Heidelberg (Lanes 12,13); S. Montevideo (Lanes 14,15); S. Dubulin (Lane 16); S. Sinstorf (Lane 17); S. salamae (Lane 18); S. Cuckmere (Lane 19); S. Othmarschen (Lane 20); S. Haifa (Lane 21); ND, not deteremind.

Table 3
Distribution of Class 1 Integron Gene Cassette and Antibiotic Resistance Patterns of 21 Integron-carrying Strains

References
1. Ridley A, Threlfall EJ. Molecular epidemiology of antibiotic resistance genes in multiresistant epidemic Salmonella typhimurium DT 104. Microb Drug Resist. 1998. 4:113–118.

2. Daly M, Buckley J, Power E, O'Hare C, Cormican M, Cryan B, Wall PG, Fanning S. Molecular characterization of Irish Salmonella enterica serotype typhimurium: detection of class I integrons and assessment of genetic relationships by DNA amplification fingerprinting. Appl Environ Microbiol. 2000. 66:614–619.

3. Guerra B, Soto S, Cal S, Mendoza MC. Antimicrobial resistance and spread of class 1 integrons among Salmonella serotypes. Antimicrob Agents Chemother. 2000. 44:2166–2169.

4. Lindstedt BA, Heir E, Nygärd I, Kapperud G. Characterization of class I integrons in clinical strains of Salmonella enterica subsp. enterica serovars Typhimurium and Enteritidis from Norwegian hospitals. J Med Microbiol. 2003. 52:141–149.

5. Rodríguez I, Rodicio MR, Herrera-León S, Echeita A, Mendoza MC. Class 1 integrons in multidrug-resistant non-typhoidal Salmonella enterica isolated in Spain between 2002 and 2004. Int J Antimicrob Agents. 2008. 32:158–164.

6. Lévesque C, Piché L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995. 39:185–191.

7. Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989. 3:1669–1683.

9. White PA, McIver CJ, Rawlinson WD. Integrons and gene cassettes in the enterobacteriaceae. Antimicrob Agents Chemother. 2001. 45:2658–2661.

10. Sallen B, Rajoharison A, Desvarenne S, Mabilat C. Molecular epidemiology of integron-associated antibiotic resistance genes in clinical isolates of enterobacteriaceae. Microb Drug Resist. 1995. 1:195–202.

11. Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995. 15:593–600.

12. Popoff MY. Antigenic formulas of the Salmonella serovars. WHO Collaborating Centre for Reference and Research on Salmonella. 2001. Paris: Institute Pasteur.
13. Anderson ES, Ward LR, Saxe MJ, de Sa JD. Bacteriophage-typing designations of Salmonella typhimurium. J Hyg (Lond). 1977. 78:297–300.
14. Choi SH, Woo JH, Lee JE, Park SJ, Choo EJ, Kwak YG, Kim MN, Choi MS, Lee NY, Lee BK, Kim NJ, Jeong JY, Ryu J, Kim YS. Increasing incidence of quinolone resistance in human non-typhoid Salmonella enterica isolates in Korea and mechanisms involved in quinolone resistance. J Antimicrob Chemother. 2005. 56:1111–1114.

15. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial suceptibility
testing; eighteenth informational supplement. 2008.
16. White PA, McIver CJ, Deng Y, Rawlinson WD. Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol Lett. 2000. 182:265–269.
17. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995. 33:2233–2239.

18. Oh YH, Song MO, Kim MS, Park SG, Lee YK. Detection of antibiotic resistant genes in Salmonella enterica Serovar Typhimurium isolated from food borne patients in Seoul using multiplex-PCR. J Bacteriol Virol. 2005. 35:183–190.
19. Sohn CK, Lee JA, Lee DO, Han W, Jung JK. Structural analysis of class I integron gene cassette and assessment of genetic relationships by PFGE of Salmonella enterica Serovar Typhimurium isolated in Gyeongbuk area. Kor J Microbiol. 2006. 42:12–18.
20. Kim TE, Kwon HJ, Cho SH, Kim S, Lee BK, Yoo HS, Park YH, Kim SJ. Molecular differentiation of common promoters in Salmonella class 1 integrons. J Microbiol Methods. 2007. 68:453–457.

21. Randall LP, Cooles SW, Osborn MK, Piddock LJ, Woodward MJ. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J Antimicrob Chemother. 2004. 53:208–216.

22. Vo AT, van Duijkeren E, Fluit AC, Wannet WJ, Verbruggen AJ, Maas HM, Gaastra W. Antibiotic resistance, integrons and Salmonella genomic island 1 among non-typhoidal Salmonella serovars in The Netherlands. Int J Antimicrob Agents. 2006. 28:172–179.

23. Cameron FH, Groot Obbink DJ, Ackerman VP, Hall RM. Nucleotide sequence of the AAD(2'') amino glycoside adenylyltransferase determinant aadB. Evolutionary relationship of this region with those surrounding aadA in R538-1 and dhfrII in R388. Nucleic Acids Res. 1986. 14:8625–8635.

24. Peters ED, Leverstein-van Hall MA, Box AT, Verhoef J, Fluit AC. Novel gene cassettes and integrons. Antimicrob Agents Chemother. 2001. 45:2961–2964.

25. Oh JY, Yu HS, Kim SK, Seol SY, Cho DT, Lee JC. Changes in patterns of antimicrobial susceptibility and integron carriage among Shigella sonnei isolates from southwestern Korea during epidemic periods. J Clin Microbiol. 2003. 41:421–423.

26. Yu HS, Lee JC, Kang HY, Ro DW, Chung JY, Jeong YS, Tae SH, Choi CH, Lee EY, Seol SY, Lee YC, Cho DT. Changes in gene cassettes of class 1 integrons among Escherichia coli isolates from urine specimens collected in Korea during the last two decades. J Clin Microbiol. 2003. 41:5429–5433.





 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download