Abstract
Background
Many genes encoding aminoglycoside modifying enzymes (AMEs) on transposon or plasmid were transferred from one strain to another strain and inserted into a staphylococcal chromosomal cassette mec (SCCmec). There are very diverse subtypes in SCCmec type to the insertion of resistant genes. Therefore, we researched the resistance rates of antibiotics and distribution of AME genes according to SCCmec type in MRSA strains.
Materials and Methods
We isolated 640 Staphylococcus aureus from non-tertiary hospitals in 2004, detected mecA, aac(6')-aph(2"), aph(3')-IIIa, and ant(4')-Ia using the multiplex PCR method, tested antibacterial susceptibility disk diffusion and minimal inhibitory concentration, and determined SCCmec type.
Results
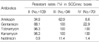
Of 640 S. aureus isolates, MRSA rate was 39.7% and all MRSA isolates carried mecA gene. Among 214 MRSA selected, aminoglycoside-resistant rates were 98.1% in kanamycin and tobramycin, 68.7% in gentamicin, 30.8% in amikacin, and 2.8% in netilmicin. The detection rates for aac(6')-aph(2"), aph(3')-IIIa, and ant(4')-Ia were 77.1%, 13.1%, and 53.3%, respectively. Also, SCCmec type was 50.9% in SCCmec type II, 16.4% in type III, and 32.7% in type IV. The genes encoding AMEs were distributed aac(6')-aph(2") (49.5%) and aac(6')-aph(2")/ant(4')-Ia (36.7%) in SCCmec type II, aph(3')-IIIa/aac(6')-aph(2") (60%) and aac(6')-aph(2") (31.4%) in type III, and aac(6')-aph(2")/ant(4')-Ia (41.4%) and ant(4')-Ia (50%) in type IV.
Figures and Tables
Table 2
Antimicrobial Resistant Rates of Staphylococcus aureus, Against Several Antibiotics from 146 Non-tertiary Hospitals

References
1. Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998. 339:520–532.
2. Fluit AC, Visser MR, Schmitz FJ. Molecular detection of antimicrobial resistance. Clin Microbiol Rev. 2001. 14:836–871.

3. Matsumura M, Karakura T, Imanaka T, Aiba S. Enzymatic and nucleotide sequence studies of a kanamycin inactivating enzyme encoded by a plasmid from thermophilic bacilli in comparison with that encoded by plasmid pUB110. J Bacteriol. 1984. 160:413–420.

4. Ubukata K, Yamashita N, Gotoh A, Konno M. Purification and characterization of aminoglycoside- modifying enzymes from Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 1984. 25:754–759.

5. Byrne ME, Rouch DA, Skurray RA. Nucleotide sequence analysis of IS256 from the Staphylococcus aureus gentamicin-tobramycin-kanamycin-resistance transposon Tn4001. Gene. 1989. 81:361–367.

6. Derbise A, Dyke KG, el Solh N. Characterization of a Staphylococcus aureus transposon, in 5405, located within in 5404 carryubg the aminoglycoside resistance genes, aphA-3 and addE. Plasmid. 1996. 35:174–188.

7. Hiramatsu K, Ito T, Hanaki H. Mechanism of methicillin and vancomycin resistance in Staphylococcus aureus. Bailliere's Clin Infect Dis. 1999. 5:221–242.
8. Préz-Roth E, Claverie-Martin F, Villar J, Mndez-lvarez S. Multiplex PCR for simultaneous identification of Staphylococcus aureus and detection of methicillin and mupirocin resistance. J Clin Microbiol. 2001. 39:4037–4041.

9. Vanhoof R, Godard C, Content J, Nyssen HJ, Hannecart-Pokorni E. Detection by polymerase chain reaction of genes encoding aminoglycoside-modifying enzymes in methicillin-resistant Staphylococcus aureus isolates of epidemic phage types. J Med Microbiol. 1994. 41:282–290.

10. Clinical and Laboratory Standards Institute. Clinical and Laboratory Standards. Performance standards for antimicrobial susceptibility testing: fifteenth informational supplement. 2005. Wayne, PA:
11. Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002. 46:2155–2161.

12. Lee H, Yong D, Lee K, Hong SG, Kim EC, Jeong SH, Park YJ, Choi TY, Uh Y, Shin JH, Lee WK, Lee J, Ahn JY, Lee SH, Woo GJ. Antimicrobial Resistance of Clinically Important Bacteria Isolated from 12 Hospitals in Korea in 2004. Korean J Clin Microbiol. 2005. 8:66–73.
13. Kim JS, Kim HS, Song W, Cho HC, Lee KM, Kim EC. Antimicrobial Resistance Profiles of Staphylococcus aureus Isolated in 13 Korean Hospitals. Kor J Lab Med. 2004. 24:223–229.
14. Kim HB, Kim T, Lee B, Kim US, Park SW, Shin J, Oh MD, Kim EC, Lee YS, Kim BS, Choe KW. Frequency of Resistance to Aminoglycoside Antibiotics in Staphylococcus aureus Isolated from Tertiary Hospitals. Infect Chemother. 2002. 34:39–46.
15. Choi SM, Kim SH, Kim HJ, Lee DG, Choi JH, Yoo JH, Kang JH, Shin WS, and Kang MW. Multiplex PCR for the Detection of Genes Encoding Aminoglycoside Modifying Enzymes and Methicillin Resistance among Staphylococcus species. J Korean Med Sci. 2003. 18:631–636.

16. Schmitz FJ, Fluit AC, Gondolf R, Beyrau R, Lindenlauf E, Verhoef J, Heinz HP, Jones ME. The prevalence of aminoglycoside resistance and corresponding resistance genes in clinical isolates of staphylococci from 19 European hospitals. J Antimicrob Chemother. 1999. 43:253–259.

17. Klingenberg C, Sundsfjord A, Rønnestad A, Mikalsen J, Gaustad P, Flægstad T. Phenotypic and genotypic aminoglycoside resistance in blood culture isolates of coagulase-negative staphylococci from a single neonatal intensive care unit, 1989-2000. J Antimicrob Chemother. 2004. 54:889–896.

18. Ardic N, Sareyyupoglu B, Ozyurt M, Haznedaroglu T, Ilga U. Investigation of aminoglycoside modifying enzyme genes in methicillin-resistant staphylococci. Microbiol Res. 2006. 161:49–54.

19. Ida T, Okamoto R, Shimauchi C, Okubo T, Kuga A. Identification of aminoglycoside-modifying enzymes by susceptibility testing: epidemiology of methicillin-resistant Staphylococcus aureus in Japan. J Clin Microbio. 2001. 39:3115–3121.

20. Wildemauwe C, Godard C, Vanhoof R, Bossuyt EV, Hannecart-Pokorni E. Change in major populations of methicillin-resistant Staphylococcus aureus in Belgium. J Hosp Infect. 1996. 34:197–203.

21. Aubry-Damon H, Legrand P, Brun-Buisson C, Astier A, Soussy CJ, Leclercq R. Reemergence of gentamicin-susceptible strains of methicillin-resistant Staphylococcus aureus: roles of an infection control program and of changes in aminoglycoside use. Clin Infect Dis. 1997. 25:647–653.

22. Ko KS, Kim YS, Song JH, Yeom JS, Lee H, Jung SI, Jeong DR, Kim SW, Chang HH, Ki HK, Moon C, Oh WS, Peck KR, Lee NY. Genotypic diversity of methicillin-resistant Staphylococcus aureus isolates in Korean hospitals. Antimicrob Agents Chemother. 2005. 49:3583–3585.

23. Ko KS, Peck KR, Sup OW, Lee NY, Hiramatsu K, Song JH. Genetic differentiation of methicillin-resistant Staphylococcus aureus strains from Korea and Japan. Microb Drug Resist. 2005. 11:279–286.
24. Tai PW, Huang CH, Lin QD, Huang YY. Molecular pattern and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolates at a teaching hospital in Northern Taiwan. J Microbiol Immunol Infect. 2006. 39:225–230.
25. Chongtrakool P, Ito T, Ma XX, Kondo Y, Trakulsomboon S, Tiensasitorn C, Jamklang M, Chavalit T, Song JH, Hiramatsu K. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin- resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob Agents Chemother. 2006. 50:1001–1012.





 PDF
PDF ePub
ePub Citation
Citation Print
Print







 XML Download
XML Download