Abstract
Due to the shortage of human organ donors for transplant, various studies of xenotransplantation, or the use of animal organs instead of human organs, have been carried out. The organs of porcine are thought to be safer and of a more suitable size for xenotransplantationthan those of nonhuman primates. Understanding the levels of expression of proteins, and their post-translational regulation, would be very practical between different species and among developing stages, though the molecular profiling for xenotransplantation has been rarely studied for porcine, while that of human and rodent is well known. Here, in this present study, we report protein regulation of the developing stages of liver (4-day old neonate, 19-day old piglet and 14-month old adult miniature pigs) using 2-DE and MALDI-TOF. From images of the three different stages, a total of 8 spotswhich were differently regulated were identified, and 5 spots were identified with MALDI-TOF MS. The data presented within this study provides critical direction relating to the development of livers of miniature pigs, which will assist future proteome analysis of the liver, and advance our understanding of the hurdles facing xenotransplantaion.
A comprehensive understanding of the regulation and range of gene expression is very important to elucidate the biological role of the encoded protein. Changes of gene expression configuration can also provide evidence concerning regulatory mechanisms, cellular functions and biochemical pathways [1]. Accordingly, research interest has focused on the identification and characterization of the functional protein products encoded by cell genome, whether independent or organized as a tissue [2].
Protein-based methods are very important because they characterize translational and post-translational modifications. Of the methods, two-dimensional electrophoresis (2DE) and matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) monitoring of proteome levels are considered to very promising tools.
The increasing shortage of human donor organ has spurred interest in the possible use of animal organs, particularly nonhuman primates, for human transplantation [3,4]. Presently, the use of nonhuman primate organs as replacements is dangerous, and their organs are too small to permit proper function in human adults [4]. Therefore, at present, most research focuses on the use of pigs instead of nonhuman primates for organ transplantation [5].
Especially, inbred miniature pigs have been introduced by a selective breeding program over the past 25 years, with the goal of successful organ xenotransplantation [4]. However, as with any xenotransplantation effort, the species barrier is an imposing and potentially dangerous obstacle [4]. The barriers to xenotransplantation involving miniature pigs include: 1) rejection due to the immunologic barrier between human and pig, 2) possibility of pathogen infection of human from pigs, 3) problems of physiologic and anatomic incompatibility among species, and 4) social agreements for response of patients.
Overcoming these issues is crucial if xenotransplantation is to be realized. To transplant pigs' organs into humans, we have to know the characteristics of proteins expressed in each organ. Protein expression in organs under the normal condition is unknown, although organs of miniature pigs are similar physiologically and anatomically with those of humans. Little is known regarding characteristics of expressed proteins according to postnatal developmental stages.
The proteome of the pig liver remains largely unexplored. Presently, we focused on the liver, with the hope of acquiring a similar level of information as has been achieved in rodents and humans. A four-day-old miniature pig (neonate),19-day-old miniature pig (piglet), and 14-month-old miniature pig (adult) were used as representative of developmental stages. The liver of each animal was analyzed by 2DE. Various protein expression spots among the three stages were chosen and the proteins were identified by MALDI-TOF MS. Unfortunately, since databases of various proteins in pigs are not as well-established asthose of humans and rodents, it was hard to identify these proteins. Still, the present findings are the first description of differentially expressed protein profiles of liver in miniature pigs according to postnatal development stage.
The miniature pigs were housed in the air barrier facility of the Center for Animal Resource Development at Seoul National University. Three pigs of different ages and developmental stages described above were used. Intramuscular anesthesia was achieved using a mixture of 2 mL ketamine ¡¤ HCl (50 mg/mL/kg), 1 mL xylazin (2.3 mg/mL/kg) and 1 mL Atropine sulfate (0.5 mg/mL/10 kg) during every painful/stressful procedure. The abdomen of each pig was shaved and disinfected with polyvinylpyrrolidone-iodine. The animals were immobilized on a surgical plane-table by adhesive ties. A median laparotomy was performed. The livers from the animals were nipped off aseptically and stored in liquid nitrate tank while being transported to the laboratory. These procedures were reviewed by the Institutional Animal and Care and Use Committee, Seoul National University, according to the university regulations.
The livers were removed immediately after sacrifice and stored at -70℃ until used. Frozen tissues (1 g) were pulverized under liquid nitrogen into a fine powder using a mortar and pestle. Protein samples were collected and homogenized in 900 µL lysis buffer (7 M urea, 2M thiourea, 2% w/v CHAPS, 2% Pharmalyte pH 3-10, 100 mMDTE). Samples were centrifuged at 50,000 rpm at 4℃ for 1 h. The supernatant was carefully removed and immediately frozen at -70℃.
2D-polyacrylamide gel electrophoresis (PAGE) was performed as previously described [6]. Aliquots containing 1 mg total protein were diluted in lysis buffer to a total volume of 450 µL. The samples were applied to a 240 mm, immobilized, nonlinear pH 3-10 IPG Drystrip (Amersham Pharmacia Biotech, Piscataway, NJ), which was rehydrated for at least 12 h. After rehydration, the strips were focused at 30 V for 3 h, 100 V for 1 h, 200 V for 1 h, 500 V for 1 h, 1,000 V for 1 h and finally at 8,000 V for 11 h to obtain approximately 90,000 Vh (Ettan™IPGphor™ II IEF systems; Amersham Pharmacia Biotech). Once isoelectric focusing was completed, the strips were equilibrated in 6M urea containing 20% v/v glycerol, 2% w/v sodium dodecyl sulfate (SDS) and 0.01% w/v full term for BPB with 10 mM Tributyl phosphine (Flukachemie, Buchs, Switzerland). SDS-PAGE was performed using and 8-18% separating gel without a stacking gel using the EttanDalt system (Amersham Pharmacia, Piscataway, NJ). The second dimension electrophoresis was carried out overnight at 3 W/gel at 20℃. The gels were stained with Coomassie G-250 (Bio-Rad Laboratories, Hercules, CA,USA).
The gels were stained with Coomassie G-250 (Bio-Rad) as previously described [7]. The stained gels were scanned using a GS 800 photometer (Bio-Rad) and analyzed with the ImageMaster™ 2D Platinum Software version 5.0 (GeneBio, Geneva, Switzerland). The digitalized 2DE gel images were compared by a matching method. Differentially expressed spots among groups (>3-fold and <1/3-fold) were analyzed and annotated.
The spots were cut into smaller pieces and digested using 12.5 ng/µL typsin (Promega, Madison, WI, USA) in 50 mM ammonium bicarbonate, pH 8.0, as previously described (Gorg et al., 2000). For MALDI-TOF MS analysis, the tryptic peptides were concentrated on a POROS 50 R2 column (Applied BioSystems, Foster City, CA, USA). After subsequent washing steps of column with 70% acetonitrile (can) in 5% FA, 100% can and 5% FA, the samples were loaded into a POROS 50 R2 column and washed with 5% FA. The sample was eluted with 2 µL of matrix solution consisting of 10 mg/mL α-cyano-4-hydroxy-cinnamic acid (Sigma-Aldrich, St. Louis, MO, USA) and then dropped onto MALDI sample plate [8].
MALDI-TOF MS was performed using the Voyager DE-PRO spectrometer (Applied Biosystems), equipped with a 337 nm nitrogen laser. The instrument was operated at an accelerating voltage of 20 kV, positive ion reflection mode, voltage grid 74.5%, guide wire voltage 0% and delay-time of 120 ns. The spectra were internally calibrated using the trypsin autolysis products (842.51 [M+H] and 2211.11 [M+H]), and searching in Swiss-Prot identified the proteins and NCBI database using Mascot (Matrix Science, London, UK). Monoisotopic peaks [MH+] were selected and all the searches were analyzed with a 50 ppm mass tolerance.
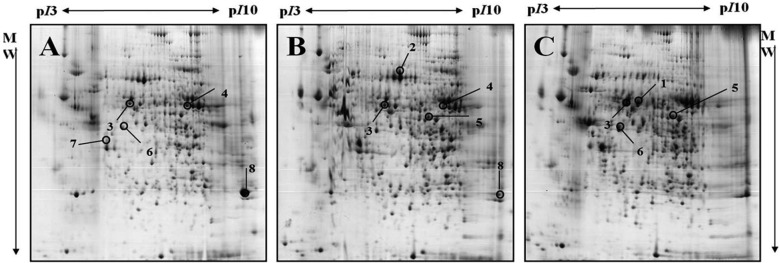
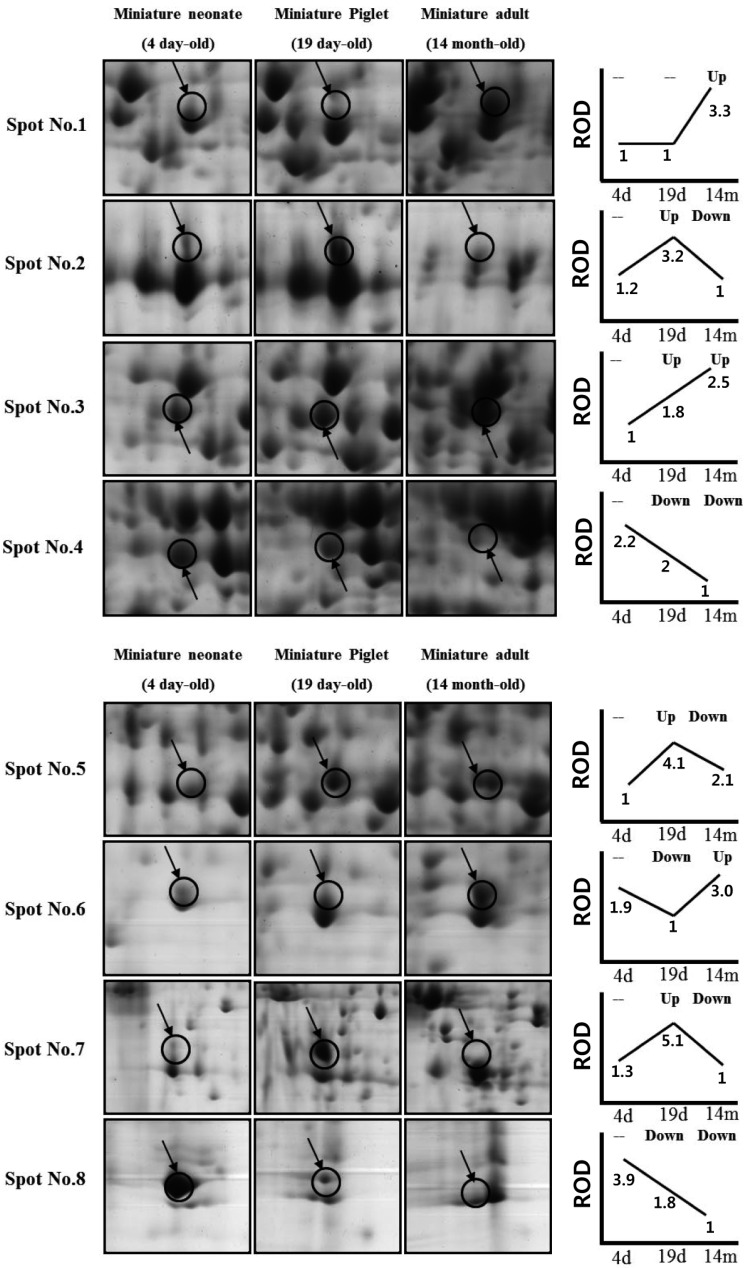
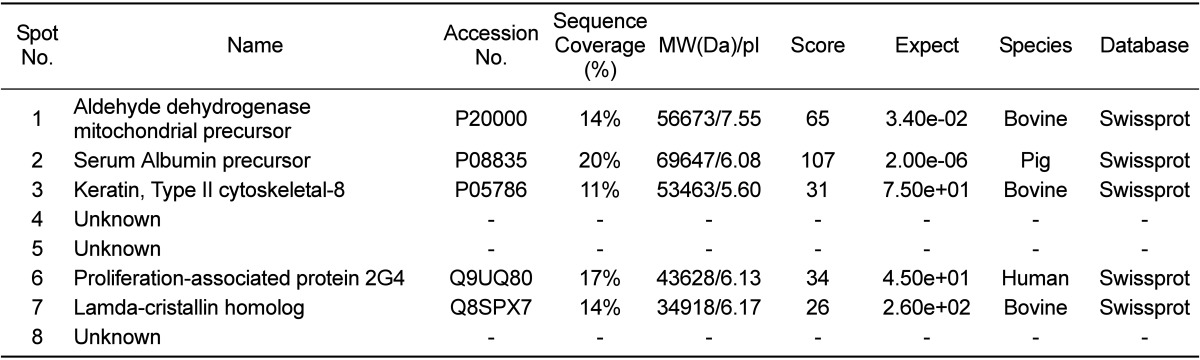
Profiles of differentially expressed liver proteins were identified with age. The liver proteins were extracted and analyzed by 2D-PAGE. After 2D gels were visualized by Coomassie blue staining, 2DE was repeated for the three pairs of matched samples, three times independently (Figure 1). For each sample, the three gels with the best resolution were selected for analysis. Analysis of spot intensities was calculated and the resulting dataset was found by Image master™ software (Geneva Bioinformatics, S.A, Geneva, Switzerland). Among the 399 protein spots detected, eight different spots were identified. The protein spots that were up-regulated>3-fold or down-regulated< 1/3-fold were selected. The selected spots were cut out from the gels and subjected to in-gel digestion with trypsin and peptide fingerprinting by MALDI-TOF MS. The peptide mass data were identified by Mascot. In this way, eight spots displayed a difference between the pigs were evident. Five spots were able to be identified by a protein function search at http://www.ebi.ac.uk/ego (Table 1).
The increase and decrease of the selected spot intensities were inspected among the three gels from neonate miniature pig to miniature adult pig (Figure 2). The expression levels of aldehyde dehydrogenase mitochondrial precursor (spot no. 1), was unchanged to up-regulated. The expression levels of serum albumin (spot no. 2) and lamda-crystallin homolog (spot no. 7) were down-regulated. Although spot no. 5 displayed a similar spot intensity patterns, correct identification could not be performed. The expression levels of keratin, a type II cytoskeletal-3 protein, were up-regulated. Proliferation-associated protein 2G4 expression was up-regulated. Although spots no. 4 and no. 8 displayed similar intensity patterns, correct identification could not be performed.
This study is, to our knowledge, the first report of protein expression of liver in miniature pig using proteome analysis according to the developmental stage. Few studies have explored the proteome of the whole liver of pigs. A comprehensive understanding of proteins expressed in the liver can provide biological information. About 400 proteins with an isoelectric point of pH 3-10 and Mr of 10-100 kDa were detected. Of 399 spots, eightprotein spots were significantly changed among groups and five spots were identified. Aldehyde dehydrogenase mitochondrial precursor belongs to the aldehyde dehydrogenase family and is associated with ethanol utilization [9]. Serum albumin is the main protein of plasma and has a good binding capacity for water Ca2+, Na+, K+, fatty acids,hormones, bilirubin and drugs. Its main function is the regulation of the colloidal osmotic pressure of blood [10]. Keratin, a type II cytoskeletal-8 protein, helps to link the contractile-apparatus to dystrophin at the costameres of striated muscle and is expressed in bladder, liver, exocervix and esophagus [11]. Proliferation-associated protein 2G4 may play a role in an ERBB3-regulated signal transduction pathway andseems to be involved in growth regulation. Itacts a co-repressorof the androgen receptor and is regulated by the ERBB3 ligandneuregulin-1/heregulin (HRG). It inhibits transcription of someE2F1-regulated promoters, probably by recruiting histone acetylase (HAT) activity. It binds RNA and associates with 28S, 18S and 5.8S maturerRNAs, several rRNA precursors and probably U3 small nucleolar RNA [12]. It may be involved in regulation of the intermediate and late steps ofrRNA processing. It may also be involved in ribosome assembly andmediatescap-independent translation of specific viral IRESs. Lamda-crystallin belongs to the 3-hydroxyacyl-CoA dehydrogenase family and is a lens component. However, it is report to be highly expressed in liver and kidney, and might be related to carcinogenesis of liver cancer [13]
Of the fiveidentified spot proteins, except for proliferation-associated protein 2G4, the proteins areexpressed in the liver. However, the identification was done with using other species' homology except serum albumin precursor. Chen et al. and Moller et al. also used human, cattle, mouse, rat homology due to shortage of database information of the pig [14,15].
Proteome analysis for xenotransplantation has been used for the investigation of target proteins for drugs and to explore the trend of protein expression after transplantation of tumor tissues into animals [16,17]. Anti-cancer drugs have been applied after transplantation of human tumor tissues into animals and the alterations of protein expression between the tumor transplanted tissue group and non-treated tissue group have been compared, with the aim of clarifying the effects of anti-cancer drugs at the protein expression level. To date, the characteristics of protein expression according to developmental stage for xenotransplantation has been essentially not studied. For xenotransplantation pigs are thought to have more potential value than non-human primates. However, one hindrance to this potential has been the paucity of mapping of the protein complement of various organs due to the scant database comparing humans and mice. At present, there are >50,000 and 25,000 database entries for humans and mice, respectively, whereas only 1072 entries for pigs were available in the MSDB database [1]. In addition, we could not confirm the proteomes (spots) identified by the 2DE and MALDI-TOF due to the lack of commercial antibodies. Knowledge of the alteration of the protein complement would provide valuable information to overcome the problems posed by the immunologic barrier, microbiologic differences and xenospecies, which present hinder xenotransplantation [3,18]. Although there is limited data for the livers of miniature pigs due to the lack of a database, the data presented in this study provides for a first reference point for the developmental livers of miniature pigs, providing critical direction for further proteome analysis of livers in future studies.
Acknowledgments
This research was supported by Bio-Industry Technology Development Program Ministry of Agriculture, Food and Rural Affairs to Prof. Je-Kyung Seong (311054-03-2-HD110) and supported by the Soonchunhyang University Fund to Prof. Sun-Shin Yi.
References
1. Junghans P, Kaehne T, Beyer M, Metges CC, Schwerin M. Dietary protein-related changes in hepatic transcription correspond to modifications in hepatic protein expression in growing pigs. J Nutr. 2004; 134(1):43–47. PMID: 14704291.

2. Pan TL, Goto S, Lord R, Huang YC, Huang CM, Wang PW, Lin YC, Kawamoto S, Ono K, Liao PC, Lin CL, Lai CY, Chang HL, Lan CH, Lee TH, Wang YC, Wu ML, Jawan B, Cheng YF, Chen ST, Chen CL. Proteome analysis in liver transplantation. Transplant Proc. 2001; 33(1-2):156. PMID: 11266756.

3. Platt JL. Physiologic barriers to xenotransplantation. Transplant Proc. 2000; 32(7):1547–1548. PMID: 11119828.

4. Cooper DK, Gollackner B, Sachs DH. Will the pig solve the transplantation backlog? Annu Rev Med. 2002; 53:133–147. PMID: 11818467.

5. Tucker A, Belcher C, Moloo B, Bell J, Mazzulli T, Humar A, Hughes A, McArdle P, Talbot A. The production of transgenic pigs for potential use in clinical xenotransplantation: baseline clinical pathology and organ size studies. Xenotransplantation. 2002; 9(3):203–208. PMID: 11983018.

6. Steiner S, Anderson NL. Pharmaceutical proteomics. Ann N Y Acad Sci. 2000; 919:48–51. PMID: 11083096.

7. Görg A, Obermaier C, Boguth G, Harder A, Scheibe B, Wildgruber R, Weiss W. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 2000; 21(6):1037–1053. PMID: 10786879.

8. Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996; 68(5):850–858. PMID: 8779443.
9. Alnouti Y, Klaassen CD. Tissue distribution, ontogeny, and regulation of aldehyde dehydrogenase (Aldh) enzymes mRNA by prototypical microsomal enzyme inducers in mice. Toxicol Sci. 2008; 101(1):51–64. PMID: 17998271.

10. Akram M, Ali Shah SM, Asif HM, Shaheen G, Shamim T, Khan MI, Ullah A, Ahmed K. Comparative study of similarity and identity of human albumin with some selected organism albumin. J Med Plants Res. 2011; 5(19):4974–4976.
11. Magin TM, Jorcano JL, Franke WW. Cytokeratin expression in simple epithelia. II. cDNA cloning and sequence characteristics of bovine cytokeratin A (no. 8). Differentiation. 1986; 30(3):254–264. PMID: 2422084.
12. Parker KA, Bruzik JP, Steitz JA. An in vitro interaction between the human U3 snRNP and 28S rRNA sequences near the alpha-sarcin site. Nucleic Acids Res. 1988; 16(22):10493–10509. PMID: 2974535.
13. Chen J, Yu L, Li D, Gao Q, Wang J, Huang X, Bi G, Wu H, Zhao S. Human CRYL1, a novel enzyme-crystallin overexpressed in liver and kidney and downregulated in 58% of liver cancer tissues from 60 Chinese patients, and four new homologs from other mammalians. Gene. 2003; 302(1-2):103–113. PMID: 12527201.

14. Chen FC, Chuang TJ. ESTviewer: a web interface for visualizing mouse, rat, cattle, pig and chicken conserved ESTs in human genes and human alternatively spliced variants. Bioinformatics. 2005; 21(10):2510–2513. PMID: 15722373.

15. Moller M, Berg F, Riquet J, Pomp D, Archibald A, Anderson S, Feve K, Zhang Y, Rothschild M, Milan D, Andersson L, Tuggle CK. High-resolution comparative mapping of pig Chromosome 4, emphasizing the FAT1 region. Mamm Genome. 2004; 15(9):717–731. PMID: 15389320.

16. Kahlem P, Dörken B, Schmitt CA. Cellular senescence in cancer treatment: friend or foe? J Clin Invest. 2004; 113(2):169–174. PMID: 14722606.

17. Besada V, Diaz M, Becker M, Ramos Y, Castellanos-Serra L, Fichtner I. Proteomics of xenografted human breast cancer indicates novel targets related to tamoxifen resistance. Proteomics. 2006; 6(3):1038–1048. PMID: 16385476.

18. Coggin JH Jr, Barsoum AL, Rohrer JW, Thurnher M, Zeis M. Contemporary definitions of tumor specific antigens, immunogens and markers as related to the adaptive responses of the cancer-bearing host. Anticancer Res. 2005; 25(3c):2345–2355. PMID: 16080461.
Figure 1
Representative 2-DE gels of livers 4-day, 19-day and 14-month old miniature pigs by developmental stages, which is visualized by Commassie blue staining. Samples of 1 mg protein were separated on pH3-10 non-linear IPG strip (24 cm) following by 8-18% gradient SDS-PAGE gels as the second dimension. Proteins were detected by Coomassie brilliant blue G-250 and compared using ImageMaster™ 2D Platinum Software version 5.0; A: 2-DE gel of 4-day old miniature pig (neonate), B: 2-DE gel of 19-day old miniature pig, C: 2-DE gel of 14-month old miniature pig. The closed circles indicate 13 differentially expressed protein spots among 4-day old miniature neonate pig, 19-day old miniature piglet, 14-month old miniature adult pig.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download