Abstract
Purpose
Allergic asthma (AA) and rheumatoid arthritis (RA) are immune tolerance-related diseases, and immune tolerance is known to be influenced by costimulatory molecules. In this study, we sought to identify common genetic susceptibility in AA and RA.
Methods
Two hundred cases of AA, 184 cases of RA, and 182 healthy controls were recruited at the Seoul National University Hospital, Seoul, Korea. Eight single nucleotide polymorphisms (SNPs) in five genes coding costimulatory molecules, namely, -318C>T, +49A>G, and 6230G>A in CTLA4, IVS3+17T>C in CD28, -3479T>G and I179V in CD86, -1C>T in CD40, and -3458A>G in CD40LG were scored, and genetic interactions were evaluated by multifactor dimensionality reduction (MDR) analysis.
Results
MDR analysis revealed a significant gene-gene interaction between -3479T>G CD86 and -3458A>G CD40LG for AA. Subjects with the T/T genotype of -3479T>G CD86 and the A/A genotype of -3458A>G CD40LG were found to be significantly more likely to develop AA than those with the T/T genotype of -3479T>G CD86 and A/- genotype of -3458A>G CD40LG (adjusted OR, 6.09; 95% CI, 2.89-12.98; logistic regression analysis controlled by age). Similarly those subjects showed a significant risk of developing RA (adjusted OR, 39.35; 95% CI, 15.01-107.00, logistic regression analysis controlled by age).
Immune tolerance refers to a specific non-reactivity to a given antigen which under other circumstances would possibly induce an immune reaction. The mechanisms underlying immune tolerance are complex and are influenced by several factors, including T cell costimulation.1 Naïve T cells require two signals to become activated.2 The first signal results from the interaction between antigen-specific T-cell receptors and an antigenic peptide presented in the context of a major histocompatibility complex class on the surface of antigen-presenting cells (APCs). The second set of signals is delivered via costimulatory molecules that are expressed on the cell surface of APCs and cytokines that are either produced by APCs, and/or by the activated T cells themselves. Costimulatory molecules belong either to the immunoglobulin family members (e.g., CD28/B7) or the tumor necrosis factor superfamily (e.g., CD40/CD40L).3
It is well known that the disruption of immune tolerance contributes to the development of allergic asthma (AA) and rheumatoid arthritis (RA). In this context, it is plausible that costimulatory molecules which initiate immune responses play important roles in the pathogeneses of these diseases.4,5 In the present study, we hypothesized that a common genetic interaction involving costimulatory molecules exists which renders susceptibility to both AA and RA. To test this hypothesis, we sought to identify genetic interactions that involved costimulatory molecules in patients with AA using multifactor dimensionality reduction (MDR) analysis, and then to confirm the relevance of such interactions in patients with RA.
Patients with AA or RA were recruited at the Seoul National University Hospital. A diagnosis of asthma was made when a subject with symptoms of dyspnea or wheezing showed reversible airway obstruction in accordance with the guidelines issued by the National Institutes of Health.6 All asthma patients showed a positive skin prick test response (allergen/histamine ratio >1.0 plus a mean wheal size >4 mm) to one or more aeroallergens. RA was diagnosed according to the 1987 revised criteria issued by the American College of Rheumatology (formerly the American Rheumatism Association).7 Control subjects were recruited from among healthy individuals. No control subjects complained of respiratory or joint symptoms as determined during interview with an allergy and rheumatology specialist, using a supporting questionnaire. In addition, they showed negative results in methacholine bronchial provocation tests (PC20≥16 mg/mL) and in skin prick tests with common aeroallergens. All subjects enrolled in this study provided written informed consent, and the study protocol was approved by the Seoul National University Hospital Institutional review board (H-1004-033-315).
Based on previous reviews,4,8 5 candidate genes encoding co-stimulatory molecules, and 8 SNPs in those genes were selected (Table 1) namely, -318C>T, +49A>G, and 6230G>A in CTLA4 (coding for cytotoxic T-lymphocyte-associated protein 4), IVS3+17T>C in CD28 (coding for CD28), -3479T>G and I179V in CD86 (coding for CD86 (B7-2)), -1C>T in CD40 (coding for CD40), and -3458A>G in CD40LG (coding for CD40 ligand). All SNPs selected were functionally relevant and previous reports showed significant associations between them and various immunologic diseases (except IVS+17T>C in CD28, see Supplemental Tables 1 and 2). Scoring was performed by the high-throughput single base-pair extension method (SNP-IT™ assay) using an SNPstream25K system, which was customized to automatically genotype DNA samples in 384-well plates and provide a colorimetric readout (Orchid Biosciences, New Jersey, USA).
Association analysis was performed for each SNP using a minor allele dominant model (i.e., AA+AB vs. BB, where A and B are major alleles, and minor alleles, respectively). P values and odds ratios (ORs) were determined by logistic regression analysis controlled for age and gender. Compliance with the Hardy-Weinberg equilibrium was determined using 2×2 tests. MDR analysis (version 3.0.2; Computational Genetics Laboratory, Dartmouth Medical School, Hanover, NH; http://www.epistasis.org) was used to reveal interactive genetic effects on AA or RA, as described previously.9 -3458A>G in CD40LG is located on the X chromosome and thus male subjects had only one allele (A/- or B/-). Because only 3 genotypes (A/A, A/B, and B/B) for one locus are allowed in MDR analysis,10 we treated A/- as A/A (or B/- as B/B). Significance of SNP combinations obtained from MDR analysis was confirmed using a contingency table and a chi-square test. All statistical analyses were performed using the R software (version 2.15.3, www.r-project.org), and P values>0.05 were regarded as significant.
A total of 200 patients with AA (age 48.4±16.7 years [mean±SD]) and 184 patients with RA (age 52.6±12.3 years [mean±SD]) participated in the study. The proportion of male was 32.0% for patients with AA and 24.5% for patients with RA, respectively. One hundred and 82 subjects were enrolled as controls (age 47.6±13.8 years [mean±SD]) and 37.4% of them were males. All eight SNPs examined were in Hardy-Weinberg equilibrium. Logistic regression analysis, adjusted for age and gender, revealed no significant association between any individual SNP and AA, and between individual SNPs and RA in a minor allele dominant model (data not shown). We then performed MDR analysis in the control and AA subjects. Table 2 summarizes the average cross validation consistency, the average prediction error, and the P value obtained by MDR analysis for each of the factors evaluated. A single two-factor model (-3479T>G CD86 and -3458A>G CD40LG) had a maximum cross validation consistency (10 out of 10) and a minimum prediction error (27.4%, P=0.001). We re-confirmed these results using a contingency table and a chi-square test. We took a simple allele counting approach to combine the results for the male and female samples instead of X chromosome analysis stratified by gender, which leads to a loss in power due to stratification.11 We considered each A/A, A/B, B/B, A/-, and B/- as separate genotypes, and then combined them. Table 3 showed combined genetic effects of -3479T>G CD86 and -3458A>G CD40LG on AA. The frequencies of each SNP combination were significantly different between the control and AA groups (df=12, chi-square=50.982, P<0.001). Subjects with a T/T genotype of -3479T>G CD86 and an A/A genotype of -3458A>G CD40LG were found to be significantly more likely to develop AA than those with a T/T genotype of -3479T>G CD86 and an A/- genotype of -3458A>G CD40LG (adjusted OR, 6.09; 95% CI, 2.89-12.98; logistic regression analysis controlled by age).
Next, we evaluated the combined effects of -3479T>G CD86 and -3458A>G CD40LG on RA (Table 4). Similarly, the frequencies of each SNP combination were significantly different between the control and RA groups (df=12, chi-square=148.925, P<0.001). In addition, we found that the T/T genotype of -3479T>G CD86 and the A/A genotype of -3458A>G CD40LG were interactively related with a significant risk of developing RA compared to a combination of a T/T genotype of -3479T>G CD86 and an A/- genotype of -3458A>G CD40LG (adjusted OR, 39.35; 95% CI 15.01-107.00; logistic regression analysis controlled by age). MDR analysis in the control and RA subjects also revealed a significant interaction between -3479T>G CD86 and -3458A>G CD40LG (Table S2).
Immune tolerance is related to conditions such as allergy, autoimmunity, tumor tolerance, and organ transplantation rejection. A previous large population-based study revealed that Th1- and Th 2-mediated diseases are significantly linked,12 which supports the theory that autoimmune and atopic diseases share risk factors that increase the propensity of the immune system to generate inappropriate responses to non-pathological antigens. AA and RA are examples of immune-tolerance-related diseases, and it is known that antigen-specific T cells play an important role in the development of these diseases. Because costimulatory signals are critical for optimal T-cell activation, proliferation, and differentiation, an enormous number of experimental and pre-clinical studies have examined the possible use of costimulatory molecules as therapeutic targets in AA and RA. Interestingly, it has been reported that RNAi-mediated CD40-CD40LG interruption not only promotes tolerance in autoimmune arthritis13 but also significantly reduces nasal allergic symptoms.14 Accordingly, genetic susceptibility involving costimulatory molecules may be a common risk factor for AA and RA.
An initial MDR analysis revealed that a genetic interaction between CD86 and CD40LG conferred susceptibility to AA and RA. CD40LG is located on the X chromosome. Most loci on the X chromosome of females are subject to X-chromosome inactivation (only one allele from each pair of alleles is expressed).15 However, a recent report raised the possibility that some loci on the X chromosome of females can escape X-chromosome inactivation.16 For example, females had higher levels of CD40LG than males16 and therefore the genetic risk to a male hemizygote (A/-) may not be equivalent to the genetic risk to a female homozygote (A/A), which requires alternate hypotheses for testing association on the X-chromosome loci.11,17 To address this, we used a simple allele counting approach to combine the results for the male and female samples, instead of X-chromosome analysis stratified by the gender, which leads to a loss in power due to stratification.11 It seems that the A/A genotype of CD40LG in a female increases the risk of AA or RA, whereas the A/- genotype of CD40LG in a male plays a protective role. However, these associations become more prominent if they are combined with the T/T genotype of -3479T>G CD86. At present, we cannot fully exclude the possibility that gender-specific traits, other than genotypes of CD40LG, may contribute to these associations.
Of the molecules involved in co-stimulatory signaling, CD40LG and its receptor CD40 have been shown to be one of the most critical. Furthermore the absence of the CD40LG-CD40 interaction results in defective T-cell-dependent immune responses.18,19 However, protective CD4 T cell responses to viral infections found in CD40- or CD40LG- deficient mice suggest the existence of an alternative pathway for priming of T cells.20,21 The up-regulation of CD86 (B7-2) on APCs has presented a CD40LG-CD40-independent pathway for T-cell priming.22 These findings suggest that the CD40LG-CD40 pathway and CD20-B7 pathway are interactively involved in the immune response. In addition, CD86 -3479T>G was significantly associated with atopy and an increased risk of asthma when combined with a promoter polymorphism (rs1800872) in the IL10.23 Although no direct interaction between CD86 and CD40LG has been reported to date, it is possible that allelic variants of the CD86 and CD40LG genes interact to promote the development of AA and RA. However, no AA patient enrolled in this study complained of arthritis symptoms, and similarly no RA patient complained of respiratory symptoms. It appears that genetic susceptibility involving costimulatory molecules contributes only to the early stages of AA or RA development, that is, the initiation of the immune response. Accordingly, AA- or RA-specific immune responses following breakdown of immune tolerance, can be initiated only in the presence of a genetic susceptibility or an environmental trigger.
SNP allele frequencies are different according to ethnicity. For example, the minor allele frequency of CD86 -3479T>G in the dbSNP database shows wide variations; 30.0% in European populations, 26.7% in Chinese populations, 21.1% in Japanese population, and 70.8% in African populations.24 In this study, the minor allele frequency of CD86 -3479T>G in the control subjects was 24.6%, which is similar to that of Chinese and Japanese populations. These data enhance the reliability of the statistical analyses performed in this study.
To evaluate the functional relevancies of CD86 -3479T>G SNP and CD40LG -3458A>G, we searched their recognition motifs in the 'RegulomeDB' database (http://regulome.stanford.edu/index). RegulomeDB includes high-throughput experimental data sets from various sources, as well as computational predictions and manual annotations to identify putative regulatory potential, and identify functional variants.25 Predicted binding motifs for -3479T>G CD86 are SRF, ELF3, TCF3, TCF7, TCF7L2, and ZFP105 and predicted binding motifs for -3458A>G CD40LG are ZFP105 and ZSCAN4, which suggests that both SNPs may be functionally competent.
Before generalizing our results, 2 points should be considered. Firstly, the case subjects enrolled in this research probably also had co-morbid diseases as well as AA or RA, which could have confounded our results. However, the inconsistent nature of co-morbid diseases, and the comparison made with apparently healthy controls might dilute any genetic effects originating from the co-morbid diseases. Secondly, we could not completely remove the risk that spurious interactions might have been identified, despite confirmation of our results by MDR analysis based on permutation. Accordingly, we suggest that our findings be confirmed by a large-scale study at a later date.
In summary, this study shows that allelic variants in the CD86 and CD40LG genes interact to significantly increase the likelihood of both AA and RA development. As mentioned above, we believe that this interaction does not entirely explain AA or RA development. Nevertheless, our results encourage us to explore genetic susceptibility conferred by costimulatory molecules in the context of immune-tolerance-related diseases.
Figures and Tables
Table 1
Genes and SNPs evaluated in this study

Table 2
Summary of MDR findings
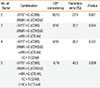
Table 3
Combined genetic effects CD86 -3479T>G and CD40LG -3458A>G on allergic asthma
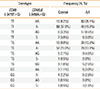
Table 4
Combined genetic effects CD86 -3479T>G and CD40LG -3458A>G on rheumatoid arthritis
ACKNOWLEDGMENTS
This study was supported by a grant from the Korea Healthcare technology R&D Project, Ministry of Health & Welfare, Republic of Korea (A070001).
References
1. Akdis M, Akdis CA. Therapeutic manipulation of immune tolerance in allergic disease. Nat Rev Drug Discov. 2009; 8:645–660.
2. Lafferty KJ, Cunningham AJ. A new analysis of allogeneic interactions. Aust J Exp Biol Med Sci. 1975; 53:27–42.
3. Chattopadhyay K, Lazar-Molnar E, Yan Q, Rubinstein R, Zhan C, Vigdorovich V, Ramagopal UA, Bonanno J, Nathenson SG, Almo SC. Sequence, structure, function, immunity: structural genomics of costimulation. Immunol Rev. 2009; 229:356–386.
4. Lombardi V, Singh AK, Akbari O. The role of costimulatory molecules in allergic disease and asthma. Int Arch Allergy Immunol. 2010; 151:179–189.
5. Goronzy JJ, Weyand CM. T-cell regulation in rheumatoid arthritis. Curr Opin Rheumatol. 2004; 16:212–217.
6. National Institutes of Health, National Heart, Lung, and Blood Institute. National Asthma Education and Prevention Program Expert Panel Report II: guidelines for the diagnosis and management of asthma. NIH Publication No. 97-4051. Bethesda (MD): U.S. Department of Health and Human Services;1997.
7. Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS, Medsger TA Jr, Mitchell DM, Neustadt DH, Pinals RS, Schaller JG, Sharp JT, Wilder RL, Hunder GG. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988; 31:315–324.
8. Peters AL, Stunz LL, Bishop GA. CD40 and autoimmunity: the dark side of a great activator. Semin Immunol. 2009; 21:293–300.
9. Motsinger AA, Ritchie MD, Reif DM. Novel methods for detecting epistasis in pharmacogenomics studies. Pharmacogenomics. 2007; 8:1229–1241.
10. Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003; 19:376–382.
11. Clayton D. Testing for association on the X chromosome. Biostatistics. 2008; 9:593–600.
12. Simpson CR, Anderson WJ, Helms PJ, Taylor MW, Watson L, Prescott GJ, Godden DJ, Barker RN. Coincidence of immune-mediated diseases driven by Th1 and Th2 subsets suggests a common aetiology. A population-based study using computerized general practice data. Clin Exp Allergy. 2002; 32:37–42.
13. Zheng X, Suzuki M, Zhang X, Ichim TE, Zhu F, Ling H, Shunnar A, Wang MH, Garcia B, Inman RD, Min WP. RNAi-mediated CD40-CD154 interruption promotes tolerance in autoimmune arthritis. Arthritis Res Ther. 2010; 12:R13.
14. Suzuki M, Zheng X, Zhang X, Ichim TE, Sun H, Kubo N, Beduhn M, Shunnar A, Garcia B, Min WP. Inhibition of allergic responses by CD40 gene silencing. Allergy. 2009; 64:387–397.
15. Chow JC, Yen Z, Ziesche SM, Brown CJ. Silencing of the mammalian X chromosome. Annu Rev Genomics Hum Genet. 2005; 6:69–92.
16. Balada E, Castro-Marrero J, Felip L, Ordi-Ros J, Vilardell-Tarrés M. Associations between the expression of epigenetically regulated genes and the expression of DNMTs and MBDs in systemic lupus erythematosus. PLoS One. 2012; 7:e45897.
17. Clayton DG. Sex chromosomes and genetic association studies. Genome Med. 2009; 1:110.
18. Kawabe T, Naka T, Yoshida K, Tanaka T, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T, Kikutani H. The immune responses in CD40-deficient mice: impaired immunoglobulin class switching and germinal center formation. Immunity. 1994; 1:167–178.
19. Grewal IS, Xu J, Flavell RA. Impairment of antigen-specific T-cell priming in mice lacking CD40 ligand. Nature. 1995; 378:617–620.
20. Oxenius A, Campbell KA, Maliszewski CR, Kishimoto T, Kikutani H, Hengartner H, Zinkernagel RM, Bachmann MF. CD40-CD40 ligand interactions are critical in T-B cooperation but not for other anti-viral CD4+ T cell functions. J Exp Med. 1996; 183:2209–2218.
21. Borrow P, Tishon A, Lee S, Xu J, Grewal IS, Oldstone MB, Flavell RA. CD40L-deficient mice show deficits in antiviral immunity and have an impaired memory CD8+ CTL response. J Exp Med. 1996; 183:2129–2142.
22. Eshima K, Choi Y, Flavell RA. CD154-CD40-independent up-regulation of B7-2 on splenic antigen-presenting cells and efficient T cell priming by staphylococcal enterotoxin A. Int Immunol. 2003; 15:817–826.
23. Bossé Y, Lemire M, Poon AH, Daley D, He JQ, Sandford A, White JH, James AL, Musk AW, Palmer LJ, Raby BA, Weiss ST, Kozyrskyj AL, Becker A, Hudson TJ, Laprise C. Asthma and genes encoding components of the vitamin D pathway. Respir Res. 2009; 10:98.
24. dbSNP. Reference SNP(refSNP) cluster report: rs2715267 [Internet]. Bethesda (MD): National Center for Biotechnology Information;2009. cited 2013 Mar 31. Available from: http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=2715267.
25. Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012; 22:1790–1797.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download