Abstract
Background
Vitiligo is an autoimmune polygenic disorder characterized by loss of pigmentation due to melanocyte destruction. The PTPN22 gene +1858 C>T single nucleotide polymorphism (rs2476601) has been shown to be associated with various autoimmune disorders.
Objective
The aim of this study was to investigate whether the PTPN22 gene +1858 C>T single nucleotide polymorphism is associated with susceptibility to generalized vitiligo in a Turkish population.
Methods
One hundred and seven patients with generalized vitiligo, and one hundred and twelve gender-, age-, and ethnic-matched controls were enrolled in the study. Genotyping was done by polymerase chain reaction-restriction fragment length polymorphism.
Vitiligo is an acquired hypomelanotic skin disorder characterized by circumscribed depigmented macules resulting from the loss of functional melanocytes from the cutaneous epidermis. Familial aggregation of vitiligo is common and usually occurs in non-Mendelian patterns, which suggests a polygenic, multifactorial mode of inheritance. Several genes such as PTPN22, MHC, CTLA4, NALP1, IL1-RN, AIS1, AIS2, AIS3, and CD4 which are also involved in autoimmunity, have been implicated in the pathogenesis of vitiligo1-3. The PTPN22 gene is located on chromosome 1p13 and encodes the lymphoid protein tyrosine phosphatase (LYP). LYP acts as a suppressor of T-cell activation and interacts with C-terminal Src kinase (CSK) which is involved in signal transduction of T-cell activation and inhibition of T-cell overactivity4-6. The PTPN22 gene 1858 C>T functional polymorphism is strongly associated with a group of autoimmune disorders including vitiligo7. The 1858 T allele of the PTPN22 gene results in hyper-responsive T cells, and patients carrying this allele have a higher risk of developing autoimmune disorders.
Controversy surrounds reports on the association between vitiligo and the PTPN22+1858 C>T polymorphism in different ethnic groups. Three separate studies conducted in English, Romanian, and English-North American populations determined an association between generalized vitiligo and the PTPN22 C>T single nucleotide polymorphism7-9. In contrast, Laddha et al.10 and Alkhateeb et al.11 found no significant association between the PTPN22 1858 C>T polymorphism and generalized vitiligo in Gujarat Indian and Jordanian populations, respectively. The objective of this study was to determine a possible association between the PTPN22 1858 C>T gene polymorphism and generalized vitiligo susceptibility in a Turkish population.
One hundred and seven patients (53 male and 54 female) who were diagnosed with generalized vitiligo and followed up in the Department of Dermatology, Medical School of Harran University were enrolled in the study. One hundred and twelve (57 male and 55 female) sex-, age-, and ethnic-matched subjects without any clinical evidence of vitiligo or any other autoimmune disorders were enrolled as control subjects. The study was approved by Harran University Medical Faculty Ethics Committee and was conducted according to the Declaration of Helsinki of 1975. All participants were informed about the nature of the study, and written informed consent was obtained.
Genomic DNA was extracted from peripheral blood leucocytes using a standard salting out procedure, as described by Miller et al.12. Polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP) assays were used to determine the +1858 C>T single nucleotide polymorphism (rs2476601) of the PTPN22gene as previously described10,13. PCR amplification was generated using the following oligonucleotide primers: forward 5'-GCCTCAATGAACTCCTCAA-3' and reverse 5'-CCTCCTGGGTTTGTACCTTA-3' (product of 400 bp).
To amplify the region containing the +1858 C>T polymorphism of exon 14 of the PTPN22 gene, the PCR reaction was carried out in a 10 µl reaction volume containing 1×PCR buffer, 2 mM MgCl2, 0.2 mM each deoxynucleotide triphosphate (dNTPs; Fermentas, St. Leon-Rot, Germany), 40 ng of DNA, 0.2 µM of each primer (Bio Basic Inc., Markham, ON, Canada), and 0.5 unit of Taq DNA Polymerase (Fermentas). The PCR conditions were: 3 minutes of initial denaturation at 94℃, followed by 30 cycles at 95℃ for 30 seconds, 30 seconds at 60℃ for annealing, and 30 seconds at 72℃ for extension, followed by 5 minutes at 72℃ for final extension. The PCR products were 400 bp.
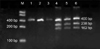
For the RFLP analysis, PCR-amplified products were digested with XcmI for 14 hours at 37℃ (New England Biolabs, Hertfordshire, UK). Anamplicon with ahomozygous TT allele of PTPN22 was cleaved by XcmI, yielding 238 and 162 bp fragments, whereas anamplicon with a homozygous CC allele remained uncut, yielding a 400 bp band. An amplicon with a heterozygous CT allele yielded three fragments of 400, 238, and 162 bp in length. The digested products were separated on a 2% agarose gel along with a 100 to 1,500 bp DNA ladder (Bio Basic Inc.). Ethidium bromide-stained gels were visualized under ultraviolet light using the Alpha Imager System (AlphaInnotech, San Leandro, CA, USA) (Fig. 1).
Data are reported as means±standard deviation, and statistical analyses were performed using the SPSS Statistical package, version 11.5 for Windows (SPSS Inc., Chicago, IL, USA). The normality of distribution was evaluated by the Kolmogorov-Smirnov test, and the normally distributed data were analyzed by Student's t-test. Genotype distributions and allele frequencies of the PTPN22 gene 1858 C>T polymorphism were analyzed with the chi-square test. All the statistical tests were two-sided, and a p-value<0.05 was considered to be statistically significant.
One hundred and seven patients with generalized vitiligo comprised of 54 females and 53 males and 112 unrelated healthy individuals comprised of 55 females and 57 males, with no autoimmune disease from the same geographical region of Turkey were enrolled in the study. The mean ages of the patients and the control groups were 27.42±14.15 and 28.38±14.41, respectively. No statistically significant difference in age or gender distribution was observed between the patient and the control groups (p>0.05). The demographic characteristics of the study group are shown in Table 1.
The genotype distributions and allele frequencies of the PTPN22 +1858 C>T polymorphism in the generalized vitiligo patients and healthy controls are shown in Table 2. The genotype frequencies of the PTPN22 +1858 C>T polymorphism were assessed in Hardy-Weinberg equilibrium in both the patients and healthy controls. The frequency of the CC and CT genotypes were 95% (102) and 5% (5) in the generalized vitiligo patients, respectively, and 94% (105) and 6% (7) in the healthy controls, respectively. There was no statistically significant difference between the generalized vitiligo patients and healthy controls according to the frequency of the heterozygote genotype (odds ratio [OR]: 0.73, 95% confidence interval [CI]: 0.22 to 2.39, p=0.60). The homozygote genotype (TT) was absent in both the generalized vitiligo patients and healthy controls. In addition, there was no significant difference between the patient and control groups with respect to allele frequencies (C or T). The frequencies of the polymorphic T allele were 2% and 3% in the patient and the control groups, respectively (OR: 0.74, 95% CI: 0.23 to 2.37, p=0.61).
Vitiligo has a polygenic and multifactorial background resulting from a complex interaction of environmental, immunological, and genetic factors. Recently, polymorphisms of a number of genes known to be involved in the development of autoimmune diseases have been implicated in susceptibility to vitiligo1-3. Since Bottini et al.14 first introduced a novel functional polymorphism in the PTPN22 gene, an increasing number of studies have linked autoimmune diseases with the PTPN22 1858 C>T polymorphism such as type I diabetes mellitus, autoimmune thyroid diseases, systemic lupus erythematosus, and rheumatoid arthritis15-17.
The PTPN22 gene encodes LYP, which is known to be involved in the control of T-cell activation. Under normal conditions, the LYP enzyme functions as a 'negative regulator' of T-cell activation and prevents overactivity of immune cells by interacting with CSK which is involved in signal transduction of T-cell activation.
The PTPN22 1858T variant results in substitution ofarginine with tryptophan, which has been shown to reduce the binding of LYP to CSK in vitro14. Decreased interaction between LYP and CSK has been suggested to inhibit the down-regulation of T-cell activation. Thus, T-cells without the LYP-CSK complex are more prone to be overactive and, consequently, more ready to evoke an autoimmune response. As a result, the PTPN22 1858 C>T polymorphism is thought to play a role in the pathogenesis of autoimmune diseases14-17.
In light of the autoimmune nature of vitiligo, Cantón et al.7 performed a study in an English population of 165 patients with generalized vitiligo and 304 healthy controls and identified an association between generalized vitiligo and the PTPN22 C>T single nucleotide polymorphism. Laberge et al.8 and LaBerge et al.9 also identified an association between generalized vitiligo and the PTPN22 gene C>T single nucleotide polymorphism in both Romanian (65 Romanian patients with generalized vitiligo and 111 control subjects) and English-North American populations (126 families with multiple cases of generalized vitiligo) However, Laddha et al.10 found no significant association between the PTPN22 1858 C>T polymorphism and generalized vitiligo in 126 Gujarat Indian patients with generalized vitiligo and 140 healthy controls. Similarly, Alkhateeb et al.11 concluded that there is no significant correlation between the PTPN22 1858 C>T polymorphism and vitiligo in a Jordanian population consisting of 55 patients with generalized vitiligo and 85 healthy controls. Our results suggest that this single nucleotide polymorphism is not associated with generalized vitiligo in Turkish patients. This was the first study to investigate the association of the PTPN22 +1858 C>T single nucleotide polymorphism in genetic susceptibility to generalized vitiligo in the Turkish population.
Previous studies have established the association between the PTPN22 1858 C>T gene polymorphism and generalized vitiligo in European and North American Caucasian populations7-9. These studies showed that the frequencies of the polymorphic (T) allele changed from 10.8% to 14.5% in vitiligo patients and from 4.1% to 8.6% in healthy controls in Caucasians. However, the results of studies of Asian populations found that the PTPN22 1858 C>T gene polymorphism is not associated with vitiligo. In our study, the allelic frequency of the mutant (T) allele in patients with vitiligo and healthy controls (2% and 3%, respectively) was lower than that in Caucasian populations. Similar to our results, in Gujarat Indian and Jordanian populations, the frequencies of the mutant (T) allele (0.79% and 2.14% in vitiligo patients and 2.14% and 2.9% in healthy controls, respectively) were lower than those of Caucasian populations (25). Furthermore, the homozygote (TT) genotype was absent in both generalized vitiligo patients and healthy controls in our study, as well as in the studies of Laddha et al.10 and Alkhateeb et al.11. These different findings on the frequencies of this polymorphism among Asian and Caucasian populations are attributed to ethnic differences.
PTPN22 may act via different pathways to evoke autoimmunity. Identification of novel genes that are associated with autoimmunity in patients with vitiligo and their exact role in the development of autoimmunity would be of great benefit. Genotyping of vitiligo patients will help identify those at higher risk of autoimmune disorders at an earlier stage of the disease. Furthermore, this would help to determine appropriate therapeutic and prophylactic approaches. Additional molecular studies with a large number of individuals from different ethnical populations are required for a better understanding of the relationship between autoimmunity and vitiligo.
Figures and Tables
Fig. 1
Polymerase chain reaction-restriction fragment length polymorphism analysis of the PTPN22 gene 1858 C/T polymorphism obtained by 2% agarose gel electrophoresis. Lane M shows 100~1,500 bp DNA ladder (Bio Basic Inc.). Lanes 1, 2 and 3 show subjects with homozygous alleles (C/C) with one intact band. Lanes 4, 5 and 6 show subjects with heterozygous alleles (C/T) showing digestion of the 400 bp product into 238 bp and 162 bp bands. No subject with homozygous allele (T/T) was observed.
References
1. Spritz RA. The genetics of generalized vitiligo and associated autoimmune diseases. Pigment Cell Res. 2007; 20:271–278.

2. Pehlivan S, Ozkinay F, Alper S, Onay H, Yuksel E, Pehlivan M, et al. Association between IL4 (-590), ACE (I)/(D), CCR5 (Delta32), CTLA4 (+49) and IL1-RN (VNTR in intron 2) gene polymorphisms and vitiligo. Eur J Dermatol. 2009; 19:126–128.

3. Zamani M, Tabatabaiefar MA, Mosayyebi S, Mashaghi A, Mansouri P. Possible association of the CD4 gene polymorphism with vitiligo in an Iranian population. Clin Exp Dermatol. 2010; 35:521–524.

4. Smyth D, Cooper JD, Collins JE, Heward JM, Franklyn JA, Howson JM, et al. Replication of an association between the lymphoid tyrosine phosphatase locus (LYP/PTPN22) with type 1 diabetes, and evidence for its role as a general autoimmunity locus. Diabetes. 2004; 53:3020–3023.

5. Hill RJ, Zozulya S, Lu YL, Ward K, Gishizky M, Jallal B. The lymphoid protein tyrosine phosphatase Lyp interacts with the adaptor molecule Grb2 and functions as a negative regulator of T-cell activation. Exp Hematol. 2002; 30:237–244.
6. Cloutier JF, Veillette A. Cooperative inhibition of T-cell antigen receptor signaling by a complex between a kinase and a phosphatase. J Exp Med. 1999; 189:111–121.

7. Cantón I, Akhtar S, Gavalas NG, Gawkrodger DJ, Blomhoff A, Watson PF, et al. A single-nucleotide polymorphism in the gene encoding lymphoid protein tyrosine phosphatase (PTPN22) confers susceptibility to generalised vitiligo. Genes Immun. 2005; 6:584–587.

8. Laberge GS, Birlea SA, Fain PR, Spritz RA. The PTPN22-1858C>T (R620W) functional polymorphism is associated with generalized vitiligo in the Romanian population. Pigment Cell Melanoma Res. 2008; 21:206–208.

9. LaBerge GS, Bennett DC, Fain PR, Spritz RA. PTPN22 is genetically associated with risk of generalized vitiligo, but CTLA4 is not. J Invest Dermatol. 2008; 128:1757–1762.

10. Laddha NC, Dwivedi M, Shajil EM, Prajapati H, Marfatia YS, Begum R. Association of PTPN22 1858C/T polymorphism with vitiligo susceptibility in Gujarat population. J Dermatol Sci. 2008; 49:260–262.

11. Alkhateeb A, Qarqaz F, Al-Sabah J, Al Rashaideh T. Clinical characteristics and PTPN22 1858C/T variant analysis in Jordanian Arab vitiligo patients. Mol Diagn Ther. 2010; 14:179–184.

12. Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988; 16:1215.

13. Majorczyk E, Jasek M, PXMLLink_XYZoski R, Wagner M, Kosior A, Pawlik A, et al. Association of PTPN22 single nucleotide polymorphism with rheumatoid arthritis but not with allergic asthma. Eur J Hum Genet. 2007; 15:1043–1048.

14. Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004; 36:337–338.

15. Peng H, Zhou M, Xu WD, Xu K, Zhai Y, Li R, et al. Association of PTPN22 C1858T polymorphism and type 1 diabetes: a meta-analysis. Immunol Invest. 2012; 41:484–496.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download