Abstract
Characteristic clinical manifestations of Newcastle disease include leukopenia and immunosuppression. Peripheral blood mononuclear cells (PBMCs) are the main targets of Newcastle disease virus (NDV) infection. To survey changes in proteomic expression in chicken PBMCs following NDV infection, PBMC proteins from 30 chickens were separated using two-dimensional electrophoresis (2-DE) and subjected to mass spectrometry analysis. Quantitative intensity analysis showed that the expression of 78 proteins increased more than two-fold. Thirty-five proteins exhibited consistent changes in expression and 13 were identified as unique proteins by matrix assisted laser desorption ionization-time of flight mass spectrometer/mass spectrometer including three that were down-regulated and 10 that were up-regulated. These proteins were sorted into five groups based on function: macromolecular biosynthesis, cytoskeleton organization, metabolism, stress responses, and signal transduction. Furthermore, Western blot analysis confirmed the down-regulation of integrin-linked kinase expression and up-regulation of lamin A production. These data provide insight into the in vivo response of target cells to NDV infection at the molecular level. Additionally, results from this study have helped elucidate the molecular pathogenesis of NDV and may facilitate the development of new antiviral therapies as well as innovative diagnostic methods.
Newcastle disease (ND) is one of the most serious disorders that affect poultry. The disease has been reported in at least six of the world's seven continents [23] and causes huge economic losses. The causative agent, Newcastle disease virus (NDV), is a member of the genus Avulavirus within the Paramyxoviridae family, and a negative-sense, single-stranded, non-segmented, and enveloped RNA virus [1,7]. The NDV genome consists of six genes that encode six structural proteins: a phosphoprotein, nucleoprotein, RNA polymerase, matrix protein, hemagglutinin-neuraminidase, and fusion protein [2]. Previous studies have shown that NDV may cause macrophages, heterophils, and peripheral blood mononuclear cells (PBMCs) to undergo apoptosis by invading lymphocytes and disrupting the monocytes-macrophage system, resulting in immunosuppression and reduced avian resistance [15,16,17].
PBMCs consist of a heterogeneous population of blood cells including monocyte and lymphocyte immune cells. The latter includes natural killer cell, B-cells, and T-cells. It is well known that PBMCs are key players in the innate and adaptive immune response because of their ability to recognize molecular patterns and evade pathogens through molecular or functional adaptation [29]. Additionally, NDV not only has a particular tropism for PBMCs but also possesses the ability to induce PBMC apoptosis [16], which is one possible explanation for immunosuppression resulting from viral infection. However, changes in protein expression and function in PBMCs following NDV infection are poorly understood.
In the current study, proteomic methods were used to characterize host peripheral blood target cell responses to NDV infection. A number of differentially expressed proteins in the PBMCs of NDV-infected chickens were identified using matrix assisted laser desorption ionizationtime of flight mass spectrometer/mass spectrometer (MALDI-TOF-MS/MS). The results presented here may provide a useful reference for future studies on the biology of chicken PBMCs and identifying proteins that influence physiological changes or immune responses associated with ND.
All animal research was conducted according to the experimental practices and standards approved (approval ID: 20111013-1) by the Animal Welfare and Research Ethics Committee at Jilin University (China).
Experimental animals for this study were 30 specific-pathogen-free chickens 6 weeks old (Harbin Veterinary Research Institute, China). The animals were allowed to adapt to the laboratory environment for several days before viral infection. Twenty chickens were inoculated intranasally with 100 µL of phosphate-buffered saline (PBS) containing 105 EID50 (50% egg infective dose) of the virulent NDV strain F48E9 (ADCC, AAS00690, USA) and designated the infection group. The other 10 chickens were inoculated intranasally with 100 µL of PBS to serve as the mock-infection group. To prevent cross-contact infection, the two groups were housed and fed separately.
Blood samples were collected at days 3 and 5 post-infection and pooled into three groups: day 3 and day 5 post-infection groups and the mock-infected group with each group including samples from 10 chickens. The blood samples were mixed with 1% EDTA as an anticoagulant in a 9:1 (v:v) ratio. After collection, the blood samples were immediately diluted in a 1:1 (v:v) ratio with PBS, layered onto the same volume of ficoll-paque (Amersham Biosciences, UK), and centrifuged at 900 × g for 20 min at room temperature. PBMCs in the middle of the interface were collected and washed with PBS. Cells isolated from each group were dissolved in lysis buffer (0.5% immobilized PH gradient [IPG] buffer, 40 mM Tris, 4% CHAPS, and 8 M urea; Roche, Switzerland), and the protein concentration was determined using a Bradford Assay kit (Amersham Biosciences, UK). Protein samples were stored at -80℃ to prevent protein degradation.
RNA was extracted from PBMCs of both groups collected on day 3 post-infection using TRIzol (Invitrogen, USA) according to the manufacturer' instructions. cDNA was prepared using a Superscript II reverse transcriptase kit (Invitrogen) and random hexamers according to the manufacturer' instructions. A pair of primers [P1 (5'-GC CATTGCYAAATACAATCC-3') and P2 (5'-GGCCCGA ATACTGTAGTCAAT-3')] was designed to amplify the partial sequence of the NDV M gene. DL2000 (Invitrogen) was used as molecular marker.
2-DE and image analysis were conducted with methods previously used in our laboratory [5]. Briefly, commercial IPG strips (pH 3~10, 24 cm; GE Healthcare, USA) were used for isoelectric focusing (IEF). The strips were rehydrated with rehydration buffer (7 M urea, 65 mM dithiothreitol [DTT], 2% CHAPS, 2 M thiourea, and 0.5% IPG buffer; GE Healthcare) that contained protein samples (1.3 mg for coomassie brilliant blue [CBB] staining or 250 µg for silver staining). After IEF, the IPG strips were equilibrated for 15 min in equilibration buffer I (6 M urea, 2% SDS, 30% glycerol, 0.002% bromophenol blue, 50 mM Tris-HCl, and 1% DTT, pH 8.8), then for 15 min in equilibration buffer II (6 M urea, 2% SDS, 30% glycerol, 0.002% bromophenol blue, 50 mM Tris-HCl, and 2.5% iodoacetamide, pH 8.8), and standard vertical SDS-PAGE (12% acrylamide) was performed for second-dimensional separation. A total of nine gels representing the three groups were processed (three replicates each for the control, samples for the day 3 post-infection group, and samples for the day 5 post-infection group). Spots in the CBB-stained gel were excised and the corresponding proteins were analyzed by a 4700 MALDI-TOF/TOF Proteomics Analyzer (Applied Biosystems, USA) following the protocol previously used in our laboratory [5]. Peptide masses were searched against the National Center for Biotechnology Information (NCBI; USA) database using the MASCOT program (Matrix Science, UK). Spots in the silver-stained gel were used for image analysis [21] that was performed using Image-Master 2D Platinum 6.0 (GE Healthcare) according to the manufacturer' protocol.
Differentially expressed proteins in the infected PBMCs were analyzed by Western blotting to confirm the 2-DE results. Two proteins, lamin A and integrin-linked kinase (ILK), were selected and analyzed based on their known functions and commercial antibodies. Briefly, equal amounts (30 µg) of protein samples from the day 5 post-infection group and mock-infected controls were subjected to separation in 12% polyacrylamide gels by SDS-PAGE. The proteins were then electrotransferred to polyvinylidene fluoride (PVDF; Millipore, USA) membranes that were subsequently blocked with 5% skim milk (Becton and Dickinson Company, USA) for 2 h at 4℃. The membranes were then incubated with mouse monoclonal antibody against lamin A (diluted 1:1000; Cell Signaling Technology, USA) or rabbit polyclonal antibody against ILK (diluted 1:1000; Cell Signaling Technology) and β-actin monoclonal antibody (diluted 1:1000; Cell Signaling Technology) for shaking overnight at 4℃. β-actin was used as a loading control. The membranes were washed with PBST (100 mM NaCl, 10 mM Tris-HCl, 0.2% Tween-20, pH 7.6) and subsequently incubated with horseradish peroxidase-conjugated anti-mouse IgG antibody (diluted 1:1000; Cell Signaling Technology) or anti-rabbit IgG antibody (diluted 1:1000; Cell Signaling Technology) for 2 h at 4℃. Antibody binding was visualized after incubation with SuperSignal West Pico Chemiluminescence Substrate (Pierce Biotechnology, USA) and photographed by manual exposure method.
All statistical analyses were performed using SPSS software (ver. 16.0, SPSS, USA). The differential expression of proteins between any two groups was analyzed using Student' t-test. Only spots corresponding to proteins with significantly different expression levels (p ≤ 0.05) with a two-fold or greater difference in intensity were selected for mass spectrometry analysis.
After intranasal inoculation with NDV strain F48E9, the chickens developed the disease within 3 days. Typical ND symptoms were observed by day 3 post-infection including anorexia, apathy, unwillingness to move around, and eyes half-open or fully closed. Blood samples were collected on days 3 and 5 post-infection, and divided into three groups as described above. PBMCs were immediately isolated from the blood samples of each group, subjected to 2-DE, and tested for NDV infection using RT-PCR. The M gene of NDV was amplified from PBMC samples of the day 3 post-infection group but not from the mock-infected group samples (Fig. 1), demonstrating that the chickens had been successfully infected.
To obtain expression profiles of the proteins, we obtained PBMC proteins from the control, day 3 post-infection, and day 5 post-infection groups for 2-DE analysis. The numbers of protein spots detected on the 24-cm 2-DE gels (pI 3-10) ranged from 679 to 781. Using the average ratios of the protein spot intensities for a comparative analysis of the 2-DE gels from the three groups, intensities for a total of 35 protein spots from NDV-infected PBMCs were found to be consistently altered (Fig. 2) including 13 corresponding to significantly down-regulated proteins (ratio of infection/control ≤ 0.5, p ≤ 0.05) and 22 significantly up-regulated proteins (ratio of infection/control ≥ 2, p ≤ 0.05).
To identify PBMC proteins that were differentially expressed, a total of 35 spots in the 2-DE gels corresponding to proteins with greater than two-fold differences in expression were excised for in-gel trypsin digestion and identified using MALDI-TOF-MS/MS. Thirteen differentially expressed proteins were successfully identified (Table 1). These proteins were sorted into five groups based on their cellular functions described in the NCBI database: macromolecular biosynthesis, cytoskeleton organization, metabolism, stress responses, and signal transduction.
In the present study, 2-DE was conducted to identify changes in PBMC protein expression patterns in chickens infected with NDV. Proteins that had expression patterns which consistently changed relative to those for the control group were selected to identify potential factors that possess specific key roles in responses to NDV. Using this approach, our quantitative analysis revealed that the expression of 78 proteins changed more than two-fold. The expression patterns for 35 changed consistently and 13 of the proteins were identified as unique by MALDI-TOF-MS/MS. Functions of the identified proteins include macromolecular biosynthesis, cytoskeleton organization, metabolism, stress responses, and signal transduction. Additionally, 2-DE results for two proteins were validated by Western blot analysis.
The expression of actin (ATCG1) was markedly increased in the infected PBMCs (Table 1). Actin is a highly conserved protein and the main component of the cytoskeleton. Any changes in actin expression could undermine the cytoskeletal network, thus inducing apoptosis [31]. Increased expression of ATCG1 that was observed (Table 1) suggests that NDV may induce apoptosis by manipulating the cytoskeletal network of the targeted cells. Actin can bind to nascent mRNA, and is involved in the anchoring, transport, and topological positioning of mRNA [24]. In infected PBMCs, actin could potentially regulate gene transcription during virus replication. Therefore, increased actin expression could be due to NDV infection to facilitate viral replication and the spreading of progeny virus particles. Data obtained in the current study strongly suggest that actin plays an important role in the NDV infection process. However, the role of changes in actin expression requires further investigation.
Four proteins related to metabolism were identified as being differentially expressed. Triosephosphate isomerase (TPI) and apolipoprotein A-I (APO-AI) were found to be up-regulated while fructose-bisphosphate aldolase C (ALDOC) and phosphoglycerate kinase (PGK) were down-regulated (Table 1). TPI is involved in glycolysis, glyceraldehyde-3-phosphate metabolism, and small-molecule metabolic processes [11]. APO-AI plays a key role in lipid transport and cholesterol metabolism [3]. Existing evidence suggests that APO-AI can prevent lymphocyte activation and proliferation by regulating the cellular cholesterol balance in peripheral lymph nodes, thus modulating immune cell function [32]. In the present study, increased APO-AI expression could be due to NDV infection, thus facilitating replication of the virus. ALDOC possesses fructose-bisphosphate aldolase activity, and is involved in glycolysis and apoptosis [22]. PGK is a glycolytic enzyme that plays an important role in cellular energy production [30]. Decreased PGK expression in the NDV-infected PBMCs suggests that the virus may reduce activity of the host glycolytic pathway. Further research is needed to identify specific changes in protein expression that are affected by NDV infection and protect the host or facilitate NDV infection.
In this study, four proteins related to transcription, RNA splicing, and protein biosynthesis were identified as being differentially expressed. The expression of heterogeneous nuclear ribonucleoprotein H3 (hnRNP H3), lamin A, PDZ and LIM domain 1 (PDLIM1), and cyclophilin A (CypA) was up-regulated (Table 1). hnRNP H3 is known to be involved in RNA splicing and participates in early heat shock-induced splicing arrest [13,34]. PDLIM1 and lamin A influence transcription while lamin A also participates in apoptosis regulation. Nuclear lamin A (a network of lamins and membrane-associated proteins) provides attachment sites for chromatin along with structural support for the nuclear membrane. It is also involved in multiple cellular functions including DNA replication and transcription [8,27]. Lamins are among the first proteins targeted during breakdown of the nuclear structure during apoptosis [18]. Therefore, we hypothesize that the purpose of up-regulated lamin A expression is to inhibit apoptosis following NDV infection.
CypA is the major binding protein of cyclosporin A in the cytosol as well as a multifunctional protein [9]. This factor is a peptidylprolyl cis-trans isomerase, and may be involved in protein folding, trafficking, assembly, immune modulation, and cell signaling due to its chaperone-like activity. CypA also plays an important role in several diseases such as viral infections, cardiovascular disease, and cancer. A recent study demonstrated that CypA possesses anti-influenza virus activity [33]. In the present study, CypA expression was up-regulated following infection by NDV, consistent with previously reported changes in CypA expression following influenza virus infection [19]. Further research is required to confirm whether CypA possesses anti-NDV activity similar to its anti-influenza activity.
It is widely known that ILK is a multi-domain signaling protein and ubiquitously expressed. ILK localizes at focal adhesions, filaments, and the centrosome, and helps regulate actin cytoskeleton organization, cell contraction, and cell adhesion. This protein is also a unique adaptor that connects cell adhesion molecules to intracellular signaling pathway factors [10]. Although no research proving whether ILK affects the replication of NDV has been performed, cumulative evidence suggests that the inhibition of ILK expression prevents Akt activation that in turn suppresses coxsackievirus B3 (CVB3) replication [6,20]. Apoptosis is an innate immune response of cells to viral infection [25]. Viruses can invade host cells and produce viral particles inside the cell; apoptosis stops virus production. However, viruses have evolved numerous strategies to prevent apoptosis [25]. One viral target within the apoptotic cascade is Akt, a downstream target of ILK that regulates cell survival and metabolism [26]. In this study, the expression of ILK was down-regulated upon infection by NDV. Therefore, it is reasonable to speculate that NDV infection led to ILK activation that triggered Akt signaling, inhibited the apoptotic pathway, and promoted replication of the virus. Further research is needed to test this hypothesis.
Apoptosis is a major factor that causes immunosuppression and leukopenia in NDV-infected chickens [15,16,17]. However, it is unclear what specific PBMC proteins are associated with apoptosis. Combined with data from previous study [16], results of the current investigation suggest that ALDOC, lamin A, ILK, and CypA may have direct or indirect effects on the apoptosis of immune cells.
In the present study, two blood coagulation factors, pre-fibrinogen alpha subunit (FGA) and pleckstrin (PLCK), were differentially expressed following NDV infection (Table 1). FGA is a component of the blood-clotting protein fibrinogen that is involved in platelet activation and blood coagulation [12]. NDV-induced differential expression of these two proteins may disrupt the hemostatic balance, leading to coagulation and thrombosis.
The last functional group included heat shock protein (HSP), which is involved in the correct folding of newly synthesized peptides. HSP expression is regulated by physiological or environment stresses [4]. HSP-90 not only exerts an anti-apoptotic effect but also plays an important role in the biosynthesis of certain membrane receptors [28]. Many viral replication-induced heat shock responses cause the relocation and up-regulated expression of HSP-90 [14]. The expression of HSP-90 was likewise up-regulated following NDV infection. We hypothesize that HSP-90 may help prevent apoptosis and aid mature viral proteins during NDV infection.
In summary, the first PBMC proteomic profile following NDV infection in chickens was obtained in the present study. During the NDV infection process, proteins found to have altered expression levels may help regulate host responses to NDV invasion by controlling cell signaling pathways and ultimately determine the course of infection. The targets and pathways of these differently expressed proteins could not be sufficiently defined, but data from this investigation provide potential guidance for further research. Additional functional studies of these proteins will not only result in a deeper understanding of the molecular pathogenesis of NDV but also assist in developing new antiviral therapies and innovative diagnostic methods.
Figures and Tables
Fig. 1
RT-PCR results for specific detection of partial Newcastle disease virus (NDV) M gene. Lane 1, infected group; Lane 2, mock-infected group; Lane M, DL2000 marker.
Fig. 2
Representative 2-dimensional electrophoresis (DE) gel images obtained for peripheral blood mononuclear cells (PBMCs) from chickens that were infected by NDV or mock-infected. (A) Mock-infected control group and (B) day 5 post-infected group. The protein spots were visualized by silver staining. MM denotes the molecular mass.
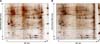
Fig. 3
Validation of altered lamin A and integrin-linked kinase (ILK) expression by Western blot analysis. (A) Magnified 2-DE images of lamin A and ILK for the mock-infected control and NDV-infected groups. (B) Western blot analysis of lamin A and ILK expression.
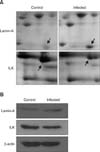
Table 1
Proteins exhibiting at least two-fold quantitative differences in expression in PBMCs from NDV-infected chickens identified by 2-DE analysis

*Spot ID represents the protein spot number on the 2-DE gels. †GI number is the MASCOT result of MALDI-TOF-MS/MS recovered from the NCBInr database. ‡Sequence coverage (%): number of amino acids spanned by the assigned peptides divided by the protein sequence length. §Matched peptides: number of peptides identified by MS/MS using MASCOT. ∥Ratio (I/U): relative protein expression in infected/uninfected samples.
References
1. Aldous EW, Alexander DJ. Newcastle disease in pheasants (Phasianus colchicus): a review. Vet J. 2008; 175:181–185.
2. Chambers P, Millar NS, Bingham RW, Emmerson PT. Molecular cloning of complementary DNA to Newcastle disease virus, and nucleotide sequence analysis of the junction between the genes encoding the haemagglutinin-neuraminidase and the large protein. J Gen Virol. 1986; 67:475–486.

3. Davidson WS, Hazlett T, Mantulin WW, Jonas A. The role of apolipoprotein AI domains in lipid binding. Proc Natl Acad Sci U S A. 1996; 93:13605–13610.
4. Didelot C, Schmitt E, Brunet M, Maingret L, Parcellier A, Garrido C. Heat shock proteins: endogenous modulators of apoptotic cell death. Handb Exp Pharmacol. 2006; 172:171–198.

5. Ding Z, Li ZJ, Zhang XD, Li YG, Liu CJ, Zhang YP, Li Y. Proteomic alteration of Marc-145 cells and PAMs after infection by porcine reproductive and respiratory syndrome virus. Vet Immunol Immunopathol. 2012; 145:206–213.

6. Esfandiarei M, Suarez A, Amaral A, Si X, Rahmani M, Dedhar S, McManus BM. Novel role for integrin-linked kinase in modulation of coxsackievirus B3 replication and virus-induced cardiomyocyte injury. Circ Res. 2006; 99:354–361.

7. Fauquet CM, Fargette D. International Committee on Taxonomy of Viruses and the 3,142 unassigned species. Virol J. 2005; 2:64.

8. Gruenbaum Y, Wilson KL, Harel A, Goldberg M, Cohen M. Review: nuclear lamins-structural proteins with fundamental functions. J Struct Biol. 2000; 129:313–323.

9. Handschumacher RE, Harding MW, Rice J, Drugge RJ, Speicher DW. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science. 1984; 226:544–547.

10. Hannigan G, Troussard AA, Dedhar S. Integrin-linked kinase: a cancer therapeutic target unique among its ILK. Nat Rev Cancer. 2005; 5:51–63.

11. Harris TK, Cole RN, Comer FI, Mildvan AS. Proton transfer in the mechanism of triosephosphate isomerase. Biochemistry. 1998; 37:16828–16838.

12. Kamath S, Lip GY. Fibrinogen: biochemistry, epidemiology and determinants. QJM. 2003; 96:711–729.

13. Kasyapa CS, Kunapuli P, Cowell JK. Mass spectroscopy identifies the splicing-associated proteins, PSF, hnRNP H3, hnRNP A2/B1, and TLS/FUS as interacting partners of the ZNF198 protein associated with rearrangement in myeloproliferative disease. Exp cell Res. 2005; 309:78–85.

14. Kobayashi K, Ohgitani E, Tanaka Y, Kita M, Imanishi J. Herpes simplex virus-induced expression of 70 kDa heat shock protein (HSP70) requires early protein synthesis but not viral DNA replication. Microbiol Immunol. 1994; 38:321–325.

15. Lam KM. Growth of Newcastle disease virus in chicken macrophages. J Comp Pathol. 1996; 115:253–263.

16. Lam KM. Newcastle disease virus-induced apoptosis in the peripheral blood mononuclear cells of chickens. J Comp Pathol. 1996; 114:63–71.

17. Lam KM, Kabbur MB, Eiserich JP. Newcastle disease virus-induced functional impairments and biochemical changes in chicken heterophils. Vet Immunol Immunopathol. 1996; 53:313–327.

18. Lazebnik YA, Takahashi A, Moir RD, Goldman RD, Poirier GG, Kaufmann SH, Earnshaw WC. Studies of the lamin proteinase reveal multiple parallel biochemical pathways during apoptotic execution. Proc Natl Acad Sci U S A. 1995; 92:9042–9046.

19. Liu N, Song W, Wang P, Lee K, Chan W, Chen H, Cai Z. Proteomics analysis of differential expression of cellular proteins in response to avian H9N2 virus infection in human cells. Proteomics. 2008; 8:1851–1858.

20. Lowenstein CJ. Integrin-linked kinase plays a key role in coxsackievirus replication. Circ Res. 2006; 99:346–347.

21. Mathesius U, Keijzers G, Natera SH, Weinman JJ, Djordjevic MA, Rolfe BG. Establishment of a root proteome reference map for the model legume Medicago truncatula using the expressed sequence tag database for peptide mass fingerprinting. Proteomics. 2006; 1:1424–1440.

22. Meighan-Mantha RL, Tolan DR. Noncoordinate changes in the steady-state mRNA expressed from aldolase A and aldolase C genes during differentiation of chicken myoblasts. J Cell Biochem. 1995; 57:423–431.

23. Miller PJ, Decanini EL, Afonso CL. Newcastle disease: evolution of genotypes and the related diagnostic challenges. Infect Genet Evol. 2010; 10:26–35.

24. Miralles F, Visa N. Actin in transcription and transcription regulation. Curr Opin Cell Biol. 2006; 18:261–266.

25. Roulston A, Marcellus RC, Branton PE. Viruses and apoptosis. Annu Rev Microbiol. 1999; 53:577–628.

26. Shiojima I, Walsh K. Role of Akt signaling in vascular homeostasis and angiogenesis. Circ Res. 2002; 90:1243–1250.

27. Stuurman N, Heins S, Aebi U. Nuclear lamins: their structure, assembly, and interactions. J Struct Biol. 1998; 122:42–66.

28. Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010; 11:515–528.

29. van der Pouw Kraan TCMT, Kasperkovitz PV, Verbeet N, Verweij CL. Genomics in the immune system. Clin Immunol. 2004; 111:175–185.

30. Wang J, Ying G, Wang J, Jung Y, Lu J, Zhu J, Pienta KJ, Taichman RS. Characterization of phosphoglycerate kinase-1 expression of stromal cells derived from tumor microenvironment in prostate cancer progression. Cancer Res. 2010; 70:471–480.

31. White SR, Williams P, Wojcik KR, Sun S, Hiemstra PS, Rabe KF, Dorscheid DR. Initiation of apoptosis by actin cytoskeletal derangement in human airway epithelial cells. Am J Respir Cell Mol Biol. 2001; 24:282–294.

32. Wilhelm AJ, Zabalawi M, Grayson JM, Weant AE, Major AS, Owen J, Bharadwaj M, Walzem R, Chan L, Oka K, Thomas MJ, Sorci-Thomas MG. Apolipoprotein A-I and its role in lymphocyte cholesterol homeostasis and autoimmunity. Arterioscler Thromb Vasc Biol. 2009; 29:843–849.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download