Abstract
Purpose
Fanconi anemia complementation group F (FANCF) is a key factor to maintaining the function of Fanconi anaemia/BRCA (FA/BRCA) pathway, a DNA-damage response pathway. However, the functional role of FANCF in breast cancer has not been elucidated. In the present study, we evaluated the chemosensitization effect of FANCF in breast cancer cells.
Methods
We performed specific knockdown of the endogenous FANCF in breast cancer cells by transfecting the cells with an FANCF short hairpin RNA (shRNA) vector. Cell viability was measured with a Cell Counting Kit-8, and DNA damage was assessed with the alkaline comet assay. The apoptosis, cell cycle, and drug accumulation were measured by flow cytometric analysis. Protein expression levels were determined by Western blot analysis, using specific antibodies.
Results
The analyses of two breast cancer cell lines (MCF-7 and MDA-MB-435S) demonstrated that the FANCF shRNA could effectively block the FA/BRCA pathway through the inhibition of Fanconi anemia complementation group D2 ubiquitination. Moreover, FANCF silencing potentiated the sensitivity of cells to mitomycin C (MMC), where combined FANCF shRNA/MMC treatment inhibited cell proliferation, induced S-phase arrest, apoptosis, and DNA fragmentation, and reduced the mitochondrial membrane potential, compared with MMC treatment alone.
Conclusion
Taken together, this study demonstrates that the inhibition of FANCF by its shRNA leads to a synergistic enhancement of MMC cytotoxicity in breast cancer cells. These results suggest that the inhibition of the FA/BRCA pathway is a useful adjunct to cytotoxic chemotherapy for the treatment of breast cancer.
Breast cancer has high morbidity and mortality rates, and is a serious threat to the health of women worldwide [1]. To date, chemotherapy has been the most frequently used treatment for breast cancer and other cancers, and has significantly prolonged the survival time of patients with breast cancer and improved their prognosis. Mitomycin C (MMC) is one of the agent for the effective treatment of metastatic breast cancer [2]. In addition, regional chemotherapy of breast cancer with MMC is regarded as a potential treatment option for unresectable recurrences and very advanced primary tumors [3]. Metabolically, the activated MMC causes DNA alkylation, monofunctional adducts formation, and DNA cross-linking that can lead to cytotoxicity and cell death [4]. Response rates of 15% to 20% are seen when MMC is used for monotherapy in the setting of metastatic breast cancer. The therapeutic value of MMC depends on the capability of cells to remove the DNA damage [5].
Fanconi anemia (FA) is an inherited chromosomal instability disorder that manifests a variety of congenital malformations, pancytopenia, and a predisposition to cancer [6]. Cells from patients with FA are particularly sensitive to DNA interstrand cross-link-inducing agents such as MMC and diepoxybutane [7]. The FA protein is a multifunctional protein composed of 15 FA complementation groups (FANC A-C, D1, D2, E, F, G, I, J, L, M, N, O, and P) [8], and is involved in the cell cycle, DNA damage and repair, apoptosis, gene transcription, and gene stability through common FA/breast cancer susceptibility gene (BRCA) cellular pathways [9]. In particular, the FA complex plays a critical role in the cell's response to chemotherapy-induced DNA damage. As an adaptor protein, Fanconi anemia complementation group F (FANCF) interacts with the complementation group C and E (FANCC/FANCE) subunits through its N-terminal, and with the complementation group A and G (FANCA/FANCG) subunits through its C-terminal. Thus, the FANCF subunit functions as the stabilizing component of the larger FA complex and maintains the biological functions of the FA/BRCA pathway [10]. FANCF inhibition, mediated by gene promoter methylation and small interfering (si) RNAs, can promote the drug sensitivity of tumor cells such as ovarian cancer [11], multiple myeloma [12], cervical cancer [13], and gliomas [14]. However, it has not been investigated whether FANCF inhibition can improve drug sensitivity in breast cancer.
In this study, we investigated the potential of FANCF silencing in enhancing the MMC tumoricidal activity in 2 well-characterized breast cancer cell lines, MCF-7 and MDA-MB-435S. MCF-7 was derived from adenocarcinoma, and MDA-MB-435S from metastatic ductal adenocarcinoma, of the breast. We hypothesized that FANCF silencing, and the subsequent dysfunction of the FA/BRCA pathway, would potentiate the antitumor activity and DNA damage caused by MMC in these representative cell lines.
The human breast cancer cell lines estrogen receptor α (ERα)-positive MCF-7 and ERα-negative MDA-MB-435S were obtained from the American Type Culture Collection (Manassas, USA). Adherent cells were maintained in Dulbecco's modified Eagle's medium, containing 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin, in a humidified atmosphere with 5% CO2 at 37℃.
Antibodies against β-actin were from Cell Signaling Technology (Beverly, USA). Antibodies against FANCF and Fanconi anemia, complementation group D2 (FANCD2) were from Abcam Inc. (Cambridge, USA). The MMC, and low melting point (LMP) and normal melting point (NMP) agarose were purchased from Sigma Chemical Co. (St. Louis, USA).
The FANCF short hairpin RNA (shRNA) expression vector was used to achieve the specific down-regulation of FANCF. In brief, DNA vectors expressing the shRNA forms were generated using the pSilencer™ 4.1-CMV plasmid (Ambion, Austin, USA). The vector expressing the FANCF shRNA oligonucleotide (5'-AACTTCCTGAAGGTGATAGCG-3') was used predominantly throughout this study. A scrambled shRNA with no significant homology to human gene sequences was used as a negative control to detect nonspecific effects.
Cells were seeded into 6-well plates (3×105 cells/well) or 100 mm dishes (2×106 cells) and allowed to adhere for 24 hours. The cells were then transfected with the pSilencer 4.1-CMV control shRNA vector (hereafter, control shRNA) or pSilencer 4.1-CMV FANCF shRNA vector (hereafter, FANCF shRNA) using Lipofectamine 2000 (Invitrogen, Carlsbad, USA), according to the manufacturer's instructions. After 4 hours, the culture medium was replaced with fresh medium supplemented with 10% FBS, and the cells were harvested at 24 and 48 hours after transfection.
Western blot analysis for the presence of specific proteins or for phosphorylated forms of proteins was performed on whole-cell sonicates and lysates of the MCF-7 and MDA-MB-435S cells. Protein (30-50 µg) was mixed in a 4:1 ratio with 5× sample buffer (20% glycerol, 4% sodium dodecyl sulfate, 10% β-mercaptoethanol, 0.05% bromophenol blue, and 1.25 M Tris-HCl [pH 6.8]; all from Sigma Chemical Co.). Equal amount of proteins were loaded onto a 10% sodium dodecyl sulfate-polyacrylamide gel. After gel electrophoresis, the cell proteins were transferred to PVDF membranes, which were then blocked with 5% milk (in Tris-buffered saline with 0.1% Tween 20) and incubated with an appropriate dilution of antibodies (1:1,000 to 1:2,000) overnight at 4℃. Thereafter, the blots were washed and incubated for 1 hour with horseradish peroxidase-conjugated anti-IgG antibody (Santa Cruz Biotechnology, Santa Cruz, USA). Immunocomplexes were visualized by using the enhanced chemiluminescence system (Santa Cruz Biotechnology, Santa Cruz, USA).
Cell viability was assessed using the Cell Count Kit-8 (Dojindo Molecular Technologies Inc., Gaithersburg, USA). Cells were seeded at 5×103 cells/well in 96-well plates and allowed to grow in the growth medium for 24 hours. The cells were then transfected with control shRNA or FANCF shRNA for 48 hours, followed by treatment with MMC at different concentrations (0.01, 0.25, 2.5, 5, 10, 20, or 40 µM) for 24 hours. Thereafter, 10 µL of CCK-8 solution was added to 100 µL of the medium in each well, and the absorbance was determined at 450 nm after 1 hour of incubation at 37℃.
Flow cytometry analysis was performed on the FACS Calibur (Becton-Dickinson, San Jose, USA). For determination of the cell cycle, 1×106 cells/well were seeded in a 6-well plate and synchronized by culturing in serum-free medium. After 48 hours, the medium was replaced with complete medium containing the desired concentrations of MMC alone, or control shRNA, or MMC plus FANCF shRNA. Floating and attached cells were collected after 24 hours of treatment and fixed in 70% ethanol overnight at 4℃. The cells were then stained with propidium iodide (PI), using PI/RNase staining buffer, for 1 hour at 37℃. Stained cells were analyzed by flow cytometry.
The cationic fluorescent carbocyanine dye, 5,5',6,6'-tetrachloro-1,1',3,3'-tetraethyl benzimidazolyl carbocyanine iodide (JC-1) was used to assess changes in the mitochondrial membrane potential (ΔΨm) observed in apoptotic cells. Cells were incubated for 15 minutes at 37℃ with 15 µg/mL JC-1 before analysis. For determination of apoptotic cells, cells were harvested, washed twice with phosphate-buffered saline (PBS), then incubated for 15 minutes at room temperature with a solution of fluorescence isothiocyanate (FITC) conjugated annexin V (2.5 µg/mL) and PI (5 µg/mL) (all from Sigma Chemical Co.), and analyzed for apoptosis.
Single-cell gel electrophoresis (comet assay) was performed, essentially according to the procedure of Singh et al. [15] with some modifications. A freshly prepared suspension of cells in 1% LMP agarose dissolved in PBS was spread onto microscope slides that had been precoated with 0.6% NMP agarose. The cells were then lysed for 1 hour at 4℃ in a buffer consisting of 2.5 M NaCl, 100 mM EDTA, 1% Triton X-100, and 10 mM Tris (pH 10). After lysis, the slides were placed in an electrophoresis unit, where the DNA was allowed to unwind for 40 minutes in the electrophoretic solution consisting of 300 mM NaOH and 1 mM EDTA (pH >13). Electrophoresis was conducted at 4℃ (the temperature of the running buffer did not exceed 12℃) for 20 minutes at 25 V and 300 mA. The slides were then neutralized with 0.4 M Tris (pH 7.5), stained with 2.5 mM PI, and covered with cover slips. To prevent additional DNA damage, all the steps described above were conducted under dimmed light or in the dark. Five hundred randomly chosen cells per slide were scanned and analyzed automatically using CometScan imaging software (Metasystems, Altlussheim, Germany). The mean tail length was calculated for about 400 cells.
Data are presented as the mean±SD, and are representative of the averages of at least 3 independent experiments. The data were analyzed using one-way analysis of variance with post-hoc analysis. A value of p<0.05 was considered statistically significant. All statistical tests were carried out by using the SPSS software version 11.5 (SPSS Inc., Chicago, USA).
ShRNA was used to knock down the FANCF expression in MCF-7 and MDA-MB-435S breast cancer cells. To verify the result of the gene silencing, FANCF expression was detected by Western blotting at 24 and 48 hours posttransfection. We found that the expressions of FANCF in the 2 cell lines were inhibited in a time-dependent manner, as compared with the control (cells treated with scrambled shRNA). The results confirmed that FANCF expression was inhibited by transfection with FANCF-targeting shRNA (Figure 1A).
FANCF is critical for maintaining the function of the FA/BRCA pathway, regulating it by maintaining the stability of FANC and FANCD2 ubiquitin activation [16]. We found that gene silencing of FANCF decreased the expression of FANCD2 and reduced its level of ubiquitination (Figure 1B). Therefore, these changes suggested that inactivation of the FA/BRCA signaling pathway was induced in the breast cancer cells. ShRNA-induced FANCF silencing led to a significant reduction of cell proliferation in both transfected cell lines in a time-dependent manner, as compared with the control cells (Figure 1C). These results further indicated that FANCF silencing blocks the function of the FA/BRCA pathway.
FANCF silencing blocked the FA/BRCA pathway, inhibiting proliferation of the breast cancer cells; the result suggested that FANCF silencing may affect the antitumor effects of MMC. The viability of nonmanipulated MCF-7 and MDA-MB-435S cells (normal FANCF expression) was markedly reduced by MMC in a concentration-dependent manner, where the estimated half maximal (50%) inhibitory concentration (IC50) values were 9.89 µM in MCF-7 cells and 10.03 µM in MDA-MB-435S cells. In contrast, the IC50 values of MMC-treated cells transfected with FANCF shRNA were significantly reduced, to 1.6 and 2.0 µM in the MCF-7 and MDA-MB-435S cells, respectively (Figure 2), representing a 83.9% and 80% decrease in IC50 of MMC, respectively. The result suggested that FANCF shRNA can synergistically improve the MMC cytotoxicity in both cell types, regardless of ERα status.
Evidence supports the view that FANCF plays an important role in DNA damage repair [17]. It has also been reported that MMC-induced DNA damage in cancer cells likely occurs at the early stage of cell proliferation inhibition [18]. Using the alkaline comet assay, we thus assessed the effect of FANCF shRNA on the MMC-induced DNA damage. MMC treatment (IC50 value) induced an increase in DNA damage, as measured by tail DNA in the comet analysis. Furthermore, the silencing of FANCF in MMC-treated cell lines led to a significant increase in the DNA damage, compared with the cells treated with MMC alone (Figure 3).
Suppression of cancer cell growth can be caused by the arrest of cell cycle progression [19]. The effect of combined FANCF shRNA/MMC treatment on the cell cycle was studied by flow cytometry, and the results are shown in Figure 4. FANCF silencing resulted in enrichment of the MMC-treated breast cancer cells in the S phase, with a concomitant decrease in the number of cells in the G0/G1 and G2/M phases. Taken together, these results showed that FANCF shRNA/MMC causes cell cycle alterations by S-phase arrest.
There is strong evidence that tumor growth is not only a result of uncontrolled proliferation but also of reduced apoptosis [20]; thus, inducing cancer cell apoptosis has been one of the key strategies in anticancer therapy. The percentage of apoptotic cells was assessed by annexin V-FITC and PI staining, followed by flow cytometric analysis. The apoptosis was detected using flow cytometry in both MCF-7 and MDA-MB-435S cell lines. It was observed that MMC increased the percentage of cells undergoing apoptosis. Interestingly, after treatment with the FANCF shRNA/MMC combination, there was a significant increase in the population of cells undergoing apoptosis, compared with the MMC-treated MCF-7 and MDA-MB-435S cells (p=0.013, p=0.008, respectively) (Figure 5).
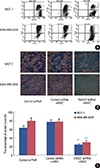
Disruption of mitochondrial integrity is one of the early events leading to apoptosis. To assess whether FANCF shRNA affects the mitochondrial function, we subsequently detected the changes of ΔΨm in treated cells. We stained cells with JC-1 dye that aggregates into healthy mitochondria and fluoresces red. After gene silencing of FANCF, we observed that there was an approximately 3-fold reduction in the ratio of red-green fluorescence intensity in MCF-7 (p=0.006) and MDA-MB-435S cells (p=0.009) treated with MMC, compared with untreated cells (Figure 6). This indicated that there was a loss of ΔΨm in FANCF shRNA-transfected cells. Taken together, all these results suggested that FANCF shRNA imparts a potent antineoplastic effect on breast cancer cells by increasing the antiproliferative effect of MMC as well as through a loss of ΔΨm, ultimately leading to apoptosis.
The FA/BRCA pathway has been reported to reverse drug resistance in some cancers, including ovarian cancer, head and neck cancer, multiple myeloma, and squamous cell lung cancer [11,12,21,22], and is therefore considered as a promising new target for therapeutic intervention. Here, as a critical factor to maintaining the biological functions of the FA/BRCA pathway, we investigated the role of FANCF in the drug sensitivity of breast cancer cells. Our studies provided evidence that FANCF chemosensitizes the breast cancer cells to MMC toxicity. Our data suggested that the inhibition of FANCF, together with MMC treatment, may provide therapeutic benefits to breast cancers. Although the chemotherapeutic agent MMC can effectively inhibit tumor growth, it is accompanied with severe side effects such as hematologic, pulmonary, and renal toxicities [23,24]; it will be of great interest to determine whether FANCF possesses the ability to reduce the effective dose of MMC, thus allowing its safe use in breast cancer treatment.
Cell cycle association with cell survival has been found in multiple human cancers. We observed that the combined treatment of breast cancer cells with MMC and FANCF shRNA resulted in S-phase arrest of cell cycle progression, suggesting that the distinct cell cycle distribution pattern contributed to the increased proliferation rate in FANCF-silenced breast cancer cells.
Deregulated growth in cancer cells is often attributed to a loss of control in the apoptotic pathways [25]. Recent studies have shown that MMC induces apoptosis by both activating caspase-3 and caspase-9 and decreasing the bcl-2 level [26]. However, it is not clear whether a combination of MMC and FANCF shRNA can synergistically provoke breast cancer cell death. In this study, we found that FANCF silencing plus MMC resulted in an increase in the apoptotic rate, compared with using MMC alone (Figure 6). Our results supported the notion that a synergistic tumor-killing effect by the FANCF-silencing/MMC treatment contributed, at least partially, to a greater effect on inducing apoptosis than MMC treatment alone.
Since the cotreatment of FANCF shRNA with MMC induced apoptosis in breast cancer cells, we wanted to know the status of the mitochondrial membrane potential in these cells. Thus, we tested the Δψm in differently treated cells by using the fluorescent dye JC-1. Upon mitochondrial collapse in apoptotic cells, JC-1 dye no longer accumulates and instead distributes throughout the cell, resulting in a decrease of red fluorescence. In accordance with this, we found that FANCF shRNA/MMC cotreatment indeed disrupted the mitochondrial membrane potential, as observed by the decreased intensity of red fluorescence. Thus, the increased apoptosis induced by FANCF shRNA/MMC cotreatment might be associated with the observed decline in Δψm.
DNA damage is one of the molecular events associated with cell cycle arrest and apoptosis. All organisms have the ability to restore genomic integrity through DNA repair. If the repair is faulty or if the cell is overwhelmed by damage, the chances are that the cell will despair and be removed by apoptosis. In our study, we found that silencing FANCF in MMC-treated cell lines led to a significant increase in DNA damage, as measured by the comet assay. We thus supposed that the cell cycle arrest and apoptosis may be attributed to the DNA damage induced by silencing FANCF.
The findings in our study, that FANCF contributes to the hypersensitization of breast cancer cells to DNA cross-linking agents such as MMC, extends the spectrum of tumor entities that may therapeutically benefit from FANCF inhibition to breast cancer. The consideration of FANCF as a potential therapeutic target, however, must take into account the part that the FA/BRCA DNA-damage repair pathway plays in maintaining genomic stability of the cell [27]. Importantly, germline or somatic inactivation of the FA/BRCA pathway has been found to result in inherited cancers or to contribute to the pathogenesis of some sporadic cancers [21,28], suggesting that FANCF inhibition could induce genetic instability and even promote carcinogenesis in some tissues. Thus, interfering with FANCF function as a therapeutic strategy bears the risk of inducing unwanted side effects and is likely to require a highly specific delivery of FANCF inhibitors to their target cells.
In conclusion, for the first time, we have shown the chemosensitizing potential of FANCF in breast cancer. FANCF silencing potentiated the MMC-induced DNA damage, S-phase cell cycle arrest, and apoptosis in MCF-7 and MDA-MB-435S cells through the activation of the intrinsic mitochondrial pathway and activation of the p38 and JNK MAPK pathways. These results suggest that FANCF could be potentially useful for the treatment of breast cancer and may serve as a chemosensitizer to improve the therapeutic efficacy of MMC, which is already in clinical use as a therapeutic drug. However, further in vitro studies, followed by animal studies and clinical trials, are necessary to validate this approach.
Figures and Tables
Figure 1
Fanconi anemia complementation group F (FANCF) levels and Fanconi anemia, complementation group D2 (FANCD2) ubiquitination in cells. MCF-7 and MDA-MB-435S cells were transfected with FANCF shRNA and control shRNA (scrambled shRNA) for 24 or 48 hours. (A) and (B) is the Western blot analysis for detecting the FANCF and FANCD2 expression changes. Then protein was extracted for performing Western blotting with anti-FANCF or anti-FANCD2 antibodies (FANCD2-L=monoubiquitinated; FANCD2-S=nonubiquitinated). β-Actin was simultaneously immunodetected to verify equal loading of cell lysates. (A) FANCF and (B) FANCD2 blots representative of three independent experiments are presented. (C) The cell viability was determined by the CCK-8 kit. *p<0.05 versus cells with control shRNA at 48 hours; †p<0.05 versus cells with control shRNA at 24 hours.

Figure 2
Changes of proliferation in mitomycin C (MMC)-treated cells. Dose-response curve of FANCF silencing in cell viability assay with or without MMC. (A) MCF-7 and (B) MDA-MB-435S cells were transfected with FANCF shRNA for 48 hours, and then treated with MMC for 24 hours. The results were expressed as a percentage relative to the control (untreated with MMC). Each point on the graph represents the means±SD.

Figure 3
DNA damage measurement. (A) Comet assay showed detectable comet tails when visualized under a fluorescent microscope, indicative of DNA damage. (B) Semiquantitation analysis of the results in (A) values of tail length (means±SD) were obtain from random selected 50 cells. *p<0.05 versus untreated cells; †p<0.05 versus cells treated with control shRNA and mitomycin C.
FANCF=Fanconi anemia complementation group F; MMC=mitomycin C.
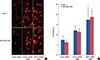
Figure 4
Cell cycle analysis. (A) MCF-7 and MDA-MB-435S cells were synchronized by culturing in serem free media for 72 hours, followed by incubation in serem-containing media for 24 hours and subsequent treatment with either mitomycin C (MMC) or MMC plus Fanconi anemia complementation group F (FANCF) shRNA or control shRNA for 24 hours. Distribution of cells in different phases of cell cycle was analyzed by propidium iodide staining followed by flow cytometry. One representative experiment is shown. Enhanced accumulation of MCF-7 and MDA-MB-435S cells in the S-phase of the cell cycle was observed after cotreatment with MMC and FANCF shRNA. (B) The bar diagrams indicate the distribution of the cells in the different phases of the cell cycle. Data are means±SD of three independent experiments in triplicates.

Figure 5
The apoptosis detection in cells. (A) Apoptosis of cells were measured using FACScan after staining with FITC-annexin V and propidium iodide. Cells in the lower right-hand quadrant are early apoptotic cells with exposed phosphatidylserine (FITC-annexin V-positive) but intact membrane (PI-negative) and (B) is the quantification of apoptosis in the indicated cell lines. *p<0.05 versus untreated cells; †p<0.05 versus cells treated with control shRNA and mitomycin C.
FANCF=Fanconi anemia complementation group F; MMC=mitomycin C; FITC=fluorescence isothiocyanate; PI=propidium iodide.

Figure 6
ΔΨm FACS detection in cells. (A) The cells were stained with 5,5',6,6'-tetrachloro-1,1',3,3'-tetraethylbe nzimidazolylcarbocyanine iodide (JC-1) fluorescence dye, and the change in ΔΨm was examined by FACS. (B) Change in ΔΨm was examined by fluorescence microscopy. (C) Densitometric analysis was done for fraction of cells. Graphs show means±SD of three independent experiments. *p<0.05 versus untreated cells; †p<0.05 versus cells treated with control shRNA and mitomycin C.
FANCF=Fanconi anemia complementation group F; MMC=mitomycin C.
Notes
This work was supported by grants from National Natural Science Foundation of China (No. 30873097, No. 30973559, and No. 81202551) and this study was also supported by Liaoning Province Doctoral Scientific Research Foundation of China (No. 20111107) and Specialized Research Fund for the Doctoral Program of Higher Education (No. 20092104110020).
References
1. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005; 55:74–108.

2. Aldabbagh K, Pouderoux S, Roca L, Poujol S, Fabbro M, Romieu G, et al. Etoposide, mitomycin, and methotrexate combination in heavily treated breast cancer: a retrospective study. Breast Cancer. 2012; 19:16–22.

3. Urruticoechea A, Archer CD, Assersohn LA, Gregory RK, Verrill M, Mendes R, et al. Mitomycin C, vinblastine and cisplatin (MVP): an active and well-tolerated salvage regimen for advanced breast cancer. Br J Cancer. 2005; 92:475–479.

4. Celli CM, Jaiswal AK. Role of GRP58 in mitomycin C-induced DNA cross-linking. Cancer Res. 2003; 63:6016–6025.
5. McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001; 2:483–490.

8. Stoepker C, Hain K, Schuster B, Hilhorst-Hofstee Y, Rooimans MA, Steltenpool J, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011; 43:138–141.

9. Bogliolo M, Lyakhovich A, Callén E, Castellà M, Cappelli E, Ramírez MJ, et al. Histone H2AX and Fanconi anemia FANCD2 function in the same pathway to maintain chromosome stability. EMBO J. 2007; 26:1340–1351.

11. Olopade OI, Wei M. FANCF methylation contributes to chemoselectivity in ovarian cancer. Cancer Cell. 2003; 3:417–420.

12. Chen Q, Van der Sluis PC, Boulware D, Hazlehurst LA, Dalton WS. The FA/BRCA pathway is involved in melphalan-induced DNA interstrand cross-link repair and accounts for melphalan resistance in multiple myeloma cells. Blood. 2005; 106:698–705.

13. Narayan G, Arias-Pulido H, Nandula SV, Basso K, Sugirtharaj DD, Vargas H, et al. Promoter hypermethylation of FANCF: disruption of Fanconi Anemia-BRCA pathway in cervical cancer. Cancer Res. 2004; 64:2994–2997.
14. Chen CC, Taniguchi T, D'Andrea A. The Fanconi anemia (FA) pathway confers glioma resistance to DNA alkylating agents. J Mol Med (Berl). 2007; 85:497–509.

15. Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988; 175:184–191.

16. Léveillé F, Blom E, Medhurst AL, Bier P, Laghmani el H, Johnson M, et al. The Fanconi anemia gene product FANCF is a flexible adaptor protein. J Biol Chem. 2004; 279:39421–39430.

17. Kowal P, Gurtan AM, Stuckert P, D'Andrea AD, Ellenberger T. Structural determinants of human FANCF protein that function in the assembly of a DNA damage signaling complex. J Biol Chem. 2007; 282:2047–2055.

18. Siegel D, Beall H, Senekowitsch C, Kasai M, Arai H, Gibson NW, et al. Bioreductive activation of mitomycin C by DT-diaphorase. Biochemistry. 1992; 31:7879–7885.

19. Gupta SC, Kim JH, Prasad S, Aggarwal BB. Regulation of survival, proliferation, invasion, angiogenesis, and metastasis of tumor cells through modulation of inflammatory pathways by nutraceuticals. Cancer Metastasis Rev. 2010; 29:405–434.

20. Tamm I, Schriever F, Dörken B. Apoptosis: implications of basic research for clinical oncology. Lancet Oncol. 2001; 2:33–42.

21. D'Andrea AD. The Fanconi Anemia/BRCA signaling pathway: disruption in cisplatin-sensitive ovarian cancers. Cell Cycle. 2003; 2:290–292.
22. Burkitt K, Ljungman M. Phenylbutyrate interferes with the Fanconi anemia and BRCA pathway and sensitizes head and neck cancer cells to cisplatin. Mol Cancer. 2008; 7:24.

23. Hortobagyi GN. Mitomycin-C in breast cancer. Semin Oncol. 1985; 12:65–70.
24. Godfrey TE. Mitomycin C in advanced breast cancer: an update. Semin Oncol. 1988; 15:71–73.
26. Jiang YY, Wang HJ, Wang J, Tashiro S, Onodera S, Ikejima T. The protective effect of silibinin against mitomycin C-induced intrinsic apoptosis in human melanoma A375-S2 cells. J Pharmacol Sci. 2009; 111:137–146.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download