Abstract
Purpose
Tumor-associated microRNAs have been detected in cancer, though whether plasma microRNA-155 (miR-155) could be a potential biomarker for laryngeal squamous cell carcinoma (LSCC) prognosis is unclear. We aimed to determine how miR-155 can be used to predict the clinical characteristics of patients with LSCC and correctly diagnose them.
Materials and Methods
We collected tissue samples and peripheral blood samples before and after treatment from 280 LSCC
cases and 560 controls. Real-time quantitative reverse transcription PCR was employed in this study to compare the relative expression of miR-155.
Results
A total of 280 LSCC patients and 560 age- and sex-matched controls were included in the study. The miR-155 level was more up-regulated in LSCC tissue than in the non-tumor tissues (13.6±2.4 vs. 3.1±0.80, p<0.001). Additionally, a significantly higher miR-155 level in plasma samples from LSCC patients than in those of the controls (8.9±1.25 vs. 1.8±0.8, p<0.001) was reported. Tissue miR-155 showed an area under the curve (AUC) of 0.933, with a sensitivity of 82.6% and a specificity of 89.2%. The AUC for plasma miR-155 was 0.757, with a sensitivity of 58.4% and a specificity of 69.5%. When early LSCC in TNM I stage was considered, tissue miR-155 showed an area under the curve of 0.804, with a sensitivity of 85.2% and a specificity of 87.3%.
Laryngeal cancer is one of the most common head and neck human cancers (excluding skin cancer) and continues to be a major unsolved health problem.1 Laryngeal squamous cell carcinoma (LSCC) is the most common histological type of laryngeal malignancy.2 It composed the most part of laryngeal cancers. Most patients of LSCC were diagnosised firstly between 50 to 75 years of age. The male-to-female sex ratio of laryngeal cancer was from 6:1 to 10:1 in most cases.3 Current methods for the diagnosis of early-stage LSCC were quite unsatisfactory.4 Nowdays, only less than 50% of patients with LSCC were in early stage at diagnosis, due to the lack of effective early-diagnosis methods.56 As effective biomarker is the key of early diagnosis of cancer and therefore the identification of new biomarkers for LSCC is in severe need.
MicroRNAs (miRNAs), an abundant class of 17-25 nucleotides that comprise small noncoding RNAs, post-transcriptionally regulate gene expression by directly binding to the 3' untranslational region (3' UTR) of target miRNAs.7 Until now, over 1000 miRNA genes have been identified in mammals; however, revealing their roles in physiology and pathology remains an ongoing process. Recently, it has been suggested that miRNAs participate in the regulation of diverse biological processes. Bioinformatics data have indicated that a single miRNA can bind to hundreds of miRNA targets, and these targets could theoretically regulate every biological process.8 Each miRNA, though powerful, has also been characterized as being marginal (a given gene can be targeted by several miRNAs, and a given miRNA typically exerts a modest repression).9 One miRNA, microRNA-155 (miR-155) is known to regulate multiple aspects of types of cancer progression in addition to demonstrating a correlation with poor prognosis.1011 Located within the noncoding B cell integration cluster (BIC) gene, miR-155 was first discovered through retroviral integrations in B cell lymphomas.12 Currently miR-155 is found to be amplified in many cancers, irrespective of retroviral integration. As one of the more highly evaluated miRNAs, miR-155 is an established oncogenic miRNA, and many of its targets are known.131415 MiR-155 is known to regulate cellular proliferation and survival through multiple mechanisms, including the mitogen activated protein kinases (MAPKs) signaling pathways. MiR-155 is a positive multi-faceted regulator of MAPK signaling, intervening at heterogeneous points along the signaling cascade to enhance signaling. Through the targeting of inositol polyphosphate-5-phosphatase (SHIP1), miR-155 enhances extracellular signal-regulated kinases (ERK) activation. In a cancer cell line, miR-155 expression enhanced ERK phosphorylation and cellular proliferation; conversely, the loss of miR-155 expression led to decreased ERK activation in B-cells.16 Downstream MAPK effectors, such as AP-1 complex member c-Jun, are enhanced through miR-155 targeting of DET1. Targeting of DET1 stabilizes c-Jun transcripts and subsequently increases AP-1 activity.17
Recently, it has been reported that plasma contains sufficiently stable miRNA species that might be useful as non-invasive biomarkers for several cancers, including LSCC.1819 Thus, the aim of the present study was to evaluate the expression of miR-155 as a potential biomarker for LSCC. However, the associations of plasma with tissue miR-155 levels are not yet clear. In this study, we investigated plasma and tissue miR-155 expression in a group of patients with LSCC and controls matched according to age and sex.
We collected tissue samples and peripheral blood samples before and after treatment from LSCC cases and controls. In total, 280 LSCC patients and 560 controls were included in this study. The clinicopathological characteristics (age, gender, smoking status, tumor size, differentiation status, lymph node metastasis, stage classification) of study subjects were recorded by trained investigators. Primary LSCC and adjacent noncancerous tissues were procured from patients undergoing surgical resections. All tissues were snap-frozen immediately after surgery and stored at -80℃ until use. Three milliliters of whole blood were collected in an Ethylenediaminetetraacetic Acid Vacutainer (BD Company, San Diego, CA, USA) for plasma. Sample collections were centrifuged at 2500 rpm at 4℃ for 10 min. The supernatant fluids were then stored at -80℃ until total RNA extraction. This study was approved by the Institutional Review Board (IRB) of the General Hospital of Tianjin Medical University, Tianjin, China.
Cell-free plasma was isolated via a two-step protocol (2500 rpm at room temperature for 10 min and 14000×g at 4℃ for 10 min) within 2 h after collection to prevent contamination of the cellular nucleic acids. The resulting plasma was transferred to new tubes and stored at -80℃. The plasma was transferred to RNase/DNase-free tubes and stored at -80℃ until RNA isolation. The total RNA was isolated from the plasma using a mirVana PARIS isolation kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions for plasma samples without enrichment for small RNAs. Briefly, 400 µL of plasma was used to extract the total RNA. Each sample was eluted in 40 µL of RNAse-free water. Cancer and non-cancer tissues from LSCC cases were immediately flash frozen in liquid nitrogen, and stored at -80℃ until RNA isolation using a mirVana PARIS isolation kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions, which were similar to those described above. The average levels of miR-155 expression in tissues and sera were normalized relative to the average amounts of U6 snRNAm, using the 2-ΔΔCT method. RNA quality was measured using a denaturing 15% polyacrylamide gel.
The reverse transcription reaction was carried out using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, America) according to the manufacturer's instructions in a total reaction volume of 7.5 mL. Quantitative polymerase chain reaction (qPCR) was performed in triplicate using TaqMan Universal PCR Master Mix (Applied Biosystems) in an ABI 7500 Real-Time PCR system (Applied Biosystems) with the following conditions: 95℃ for 10 min, followed by 40 cycles of 95℃ for 15 s and 60℃ for 1 min. The cycle threshold (Ct) values were calculated using SDS 2.0.1 software (Applied Biosystems). Template-free controls for both RT and PCR were included in each experiment to ensure target-specific amplification. In addition, if CtU6 did not occur within 32 cycles, the assay was repeated. Samples with low U6 snRNA levels were not excluded from data analyses. Primers used in qPCR are presented in Table 1.
We used the paired Wilcoxon nonparametric test to analyze pairs of non-tumor tissue and cancer samples or between cases and controls. The Mann-Whitney U test was used for the analyses of the expression of different miRNAs. Receiver operating characteristic (ROC) curves were drawn, and the areas under the ROC curves (AUCs) were measured to assess the specificity and sensitivity of circulating miR-155 as potential diagnostic biomarkers for LSCC. All statistical analyses were performed using SPSS 16.0 software (SPSS Inc., Chicago, IL, USA). Differences were considered significant if p <0.05. All experiments were performed in triplicate and repeated at least three times.
A total of 280 LSCC patients and 560 age, sex-matched controls were included in the study. The median age of the patients was 64.8±12.1 years, while the median age of the controls was 65.1±9.8 years. No differences were detected between the groups (p=0.162). Of the 280 patients, 245 were female, and the others were male. Among the 560 controls, 245 were female, and the others were male (p=0.541). However, when tobacco smoking status was considered, the rate of smoking was higher in patients with LSCC (p<0.001). The details regarding clinicopathologic features were only collected for the patients, who also came for follow-up treatment. Pathological features were obtained for all cases, among which 243 were in the 0-3-cm group while 37 were in the >3-cm group. Most patients had a tumor of more than stage I (211 of 288, 75.36%). A moderately or well differentiated tumor was seen in 197 cases, while poor differentiated tumors comprised 24.64% of the patient population. Of the patients with lymph node metastasis information, 167 had negative lymph node metastasis, and 116 had positive lymph node metastasis (Table 2).
Total RNA was isolated from the cancer and control tissues of all LSCC cancer patients. Additionally, plasma miR-155 levels were detected in both cases and controls. In general, the miR-155 levels were quantified using a quantitative reverse transcription PCR assay with U6 as the internal control. In our results, we found that the miR-155 level was more up-regulated in LSCC tissue than in the non-tumor tissues (13.6±2.4 vs. 3.1±0.80; p<0.001). Moreover, we observed a significantly higher miR-155 level in plasma samples from LSCC patients than in those from the controls (8.9±1.25 vs. 1.8±0.8; p<0.001). We also reported the miR-155 levels of the LSCC cases for each different stage. It was found that both tissue and plasma miR-155 levels were higher in stage I LSCC cases. Additionally, the levels of miR-155 in stage II, III, and IV were higher than that of stage I (Table 3). However, no increasing trend was observed as the stage increased. The expression data for miR-155 in tissue and plasma are presented in Fig. 1.
We also conducted advanced analyses to detect a possible relationship between the tissue and plasma levels of miR-155. Both tissue and plasma samples were used in the detection of the miR-155 level. On Pearson correlation analyses, a significant association was detected (p<0.001; r=0.725) (Fig. 2).
The diagnostic performances of tissue and plasma miR-155 for LSCC were detected. Cutoff points were determined using the highest sum of sensitivity and specificity. The cutoff points for tissue and plasma miR-155 were 7.34 and 3.12, respectively. Tissue miR-155 showed an AUC of 0.933, with a sensitivity of 82.6% and a specificity of 89.2% (Fig. 3A). However, the AUC for plasma miR-155 was 0.757, with a sensitivity of 58.4% and a specificity of 69.5% (Fig. 3B).
We also conducted advanced analyses on the diagnostic performances for early TNM I stage LSCC. As well, the cutoff points were determined using the highest sum of sensitivity and specificity. The cutoff points for tissue and plasma miR-155 were 7.34 and 3.12, respectively. Tissue miR-155 showed an AUC of 0.804, with a sensitivity of 85.2% and a specificity of 87.3% (Fig. 4A). However, the AUC for plasma miR-155 was 0.824, with a sensitivity of 72.4% and a specificity of 76.9% (Fig. 4B).
A total of 280 LSCC cases were obtained to detect the change of plasma miR-155. Compared to preoperative expression, plasma miR-155 demonstrated significant depression. As shown in Fig. 5, the expression of tissue miR-155 changed from 8.9 to 4.3 (p=0.002). In the smoking group, the expression of plasma miR-155 was 11.5±1.5, while the postoperative level was 5.2±0.9 (p=0.001). The level of miR-155 in the non-smoking group changed from 8.1±0.9 to 4.0±1.3, and a significant difference was detected (p=0.012).
Sensitive and specific biomarkers were capital in developing preventive screening kinds of cancers, including LSCC. However, current methods were insufficient in detecting LSCC in the early stage. Recent achievements in imaging technology have greatly improved the diagnosis of LSCC; however, the high cost maked it unavailable in developing countries. In recent years, miRNAs have been regarded as new diagnostic tools for the detection of kinds of cancers. As miRNA expression was tissue-specific and quite stable, miRNAs may be used as a marker to resolve tumor tissue from normal tissue or as a diagnostic tool. Although several groups have investigated the potential application of miRNA as a biomarker for LSCC,2021 most of the reports have used histological specimens and thus have limited clinical significance. Given that miRNAs are stable in circulation, we propose that they may serve as ideal noninvasive biomarkers for the diagnosis of LSCC. Therefore, we analyzed both tissue and plasma levels of miR-155 in patients with LSCC and controls and evaluated their diagnostic performance as diagnostic markers of LSCC.
To our knowledge, this is the first study to explore the diagnostic impact of miR-155 in a case-control study with 500 participants. Previous reports indicated that miR-155 was up-regulated in several malignancies, such as Hodgkin lymphoma,22 breast cancer,23 and lung carcinoma,24 and perceived an oncogene, although the diagnostic impact was not addressed in these studies. Similarly, in LSCC, miR-155 has been interpreted as an oncogene. In a global miRNA profiling study with 51 formalin-fixed archival LSCC samples using a quantitative reverse transcription-PCR approach, miRNA expression was correlated with patients' clinical parameters. Thirty-eight of 117 (33%) consistently detected miRNAs were significantly differentially expressed in malignant tissues when compared to normal tissues, and overexpression of miR-155 in LSCC patients was detected.25
As in most previous studies, the diagnostic effects of miR-155 on different cancers were analyzed in the tissues.2627 It has been reported that serum miRNAs were quite stable and could hardly degraded by enzymes. It has also been demonstrated that miRNAs are resistant to cancers. However, as the importance of circling miRNAs is considered in the diagnosis of cancer, the application of plasma or serum miR-155 in the diagnosis of breast cancer,28 glioma,29 lung adenocarcinoma,24 and Hodgkin lymphoma.22 In a previous study, the levels of miR-155 in cancer tissues or cell lines were significantly higher than in benign tissue or non-cancer cells.30 However, miR-155 levels in plasma were not reported in previous studies. In our study, we conducted a case-control study to identify the expression and association of both tissue and plasma miR-155 in LSCC cases. In general, our data showed that the expressions of miR-155 in both tissue and plasma were significantly higher. Moreover, we found that the expression levels of miR-155 in tissue and plasma were significantly associated. This result was accordant with several previous studies, such as microRNA-375 in colorectal cancer31 and miR-106b~25 expressions in patients with gastric cancers.32 MiR-155 in tissue and plasma has been used as a diagnostic marker for LSCC. According to the requirements for biomarkers in diseases, we speculated that both tissue and plasma miR-155 could be used as diagnostic markers for LSCC. Therefore, all of the data in our study were sufficient for describing diagnostic markers for the diagnosis of LSCC in all stages. Our study was the first to detect both tissue and plasma miR-155 in LSCC patients by using qPCR. This technology could be used to discover new biomarkers for LSCC from 280 LSCC cases and 560 controls with reasonable reliability. In the current study, we analyzed the expression of miR-155 in LSCC tissues and plasma samples in order to determine their potential value in the diagnosis of LSCC patients and early-stage LSCC. Our data showed that levels of tissue and plasma miR-155 expression were more up-regulated in LSCC than in the corresponding non-cancerous laryngeal tissues or the controls, and the ROC data showed that the AUC for tissue and plasma miR-155 was 0.933 and 0.757, respectively. In stage I LSCC patients, the ROC data indicated that the AUC for tissue and plasma miR-155 was 0.804 and 0824, respectively. Based on these ROC analyses, we found that the tissue and plasma miR-155 levels were potential important diagnostic markers for LSCC. A total of 280 LSCC cases were obtained to detect the change of plasma miR-155. Compared to the preoperative expression, plasma miR-155 demonstrated significant depression. The changes in plasma miR-155 demonstrated the potential application of miR-155 for the prognosis of LSCC. In this paper, we found that only plasma miR-155 was differently expressed in the LSCC cases. It is an interesting result that suggests that miR-155 level was more related with the pathological characteristics. When the plasma miR-155 level was considered, the expression of circulating miR-155 was modified after tobacco smoke exposure. In a study conducted by Herberth, et al.,33 it was reported that blood miRNAs are sensitive to environmental stressors, including tobacco smoke. In conclusion, we found that plasma miRNA was sensitive to different clinical and environmental factors.
The tumor promoter roles of miR-155 in different cancers are determined by both in vitro and in vivo study, and an up-regulated level of miR-155 expression would increase cell viability, prevent cell apoptosis, and increase tumorigenicity. As an important regulatory molecule, miR-155 played a key role in kidns of oncogenic processes. It could down-regulate B-cell lymphoma 6 (BCL6) protein, which could modulate the interleukin 4 responses of B cells.34 Additionally, miR-155 could act to enhance transcription and contribute to the pathogenesis of leukemia. MiR-155 could reduce the expression of BCL6 and then leads to up-regulation of certain BCL6 targets, including inhibitor of differentiation (Id2), interleukin-6 (IL6), cMyc, and Mip1α/Ccl3, all of which promote cell survival and proliferation.35 In an in vitro and in vivo study, it was found that the expression levels of miR-155 in LSCC were significantly higher than those in the control mucosa tissues.36 Down-regulation of SOCS1 expression and elevated expression of STAT3 were also observed in LSCC. The relevance of the three factors was statistically significant. Moreover, knockdown of miR-155 elevated SOCS1 expression level, suppressed STAT3 expression, and inhibited hep-2 cell growth, migration, and invasion, whereas overexpression of miR-155 resulted in the opposite effects. STAT3 protein level in poor or moderate cell differentiation was significantly higher than that in a higher degree of cell differentiation. These data demonstrated the aberrant expression and function of miR-155 and its downstream targets in LSCC. These findings suggest that miR-155 plays a promoting role during the development of LSCC, and it may be a useful marker for the prognosis and assessment of therapeutic effects.30
In conclusion, the expressions of tissue and plasma miR-155 were significantly up-regulated in patients with LSCC. Due to its reasonable sensitivity and specificity for LSCC, tissue and plasma miR-155 could serve as potential biomarkers for determining both LSCC stage and early cases. Our work will serve as a basis for further investigation, preferably large-scale validation in clinical trials, before plasma miR-155 can be used as a routine screening tool for LSCC.
Figures and Tables
Fig. 1
Relative expression of tissue and plasma miR-155 in LSCC patients. miR-155, microRNA-155; LSCC, laryngeal squamous cell carcinoma.

Fig. 2
The association between tissue and plasma miR-155 in LSCC cases. miR-155, microRNA-155; LSCC, laryngeal squamous cell carcinoma.

Fig. 3
Receiver operating characteristic curves of tissue and plasma miR-155 for the diagnosis of LSCC. (A) Tissue miR-155. (B) Plasma miR-155. AUC, area under the curve; miR-155, microRNA-155; LSCC, laryngeal squamous cell carcinoma.

Fig. 4
Receiver operating characteristic curves of tissue and plasma miR-155 for the diagnosis of early-stage LSCC. (A) Tissue miR-155. (B) Plasma miR-155. AUC, area under the curve; miR-155, microRNA-155; LSCC, laryngeal squamous cell carcinoma.

Fig. 5
Preoperative and postoperative plasma miR-155 levels of LSCC patients. miR-155, microRNA-155; LSCC, laryngeal squamous cell carcinoma.

Table 1
Primers Used in qPCR

| MiRNAs | Primer sequence (5'-3') | |
|---|---|---|
| MiR-155 | Forward | TGCCTCCAACTGACTCCTAC |
| Reverse | GCGAGCACAGAATAATACGTA | |
| U6 | Forward | GCTTCGGCAGCACATATACTAAAAT |
| Reverse | CGCTTCACGAATTTGCGTGTCAT |
Table 2
Clinicopathological Features of 280 LSCC Patients and 560 Controls
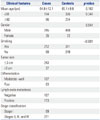
Table 3
Associations of Aberrant MiR-155 Expression in Tissue and Plasma with Clinicopathological Data of LSCC Patients

References
1. Gourin CG, Starmer HM, Herbert RJ, Frick KD, Forastiere AA, Eisele DW, et al. Short- and long-term outcomes of laryngeal cancer care in the elderly. Laryngoscope. 2015; 125:924–933.

2. Landry D, Glastonbury CM. Squamous cell carcinoma of the upper aerodigestive tract: a review. Radiol Clin North Am. 2015; 53:81–97.
3. Rutt AL, Hawkshaw MJ, Sataloff RT. Laryngeal cancer in patients younger than 30 years: a review of 99 cases. Ear Nose Throat J. 2010; 89:189–192.

4. Park JM, Jung CK, Choi YJ, Lee KY, Kang JH, Kim MS, et al. The use of an immunohistochemical diagnostic panel to determine the primary site of cervical lymph node metastases of occult squamous cell carcinoma. Hum Pathol. 2010; 41:431–437.

5. Qiu G, Li Y, Liu Z, Wang M, Ge J, Bai X. Clinical value of serum HMGB1 in diagnosis and prognosis of laryngeal squamous cell carcinoma. Med Oncol. 2014; 31:316.

6. Vasca V, Vasca E, Freiman P, Marian D, Luce A, Mesolella M, et al. Keratin 5 expression in squamocellular carcinoma of the head and neck. Oncol Lett. 2014; 8:2501–2504.

7. Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015; 518:107–110.

8. Loh YH, Yi SV, Streelman JT. Evolution of microRNAs and the diversification of species. Genome Biol Evol. 2011; 3:55–65.

9. Ceppi P, Peter ME. MicroRNAs regulate both epithelial-to-mesenchymal transition and cancer stem cells. Oncogene. 2014; 33:269–278.

10. Xu TP, Zhu CH, Zhang J, Xia R, Wu FL, Han L, et al. MicroRNA-155 expression has prognostic value in patients with non-small cell lung cancer and digestive system carcinomas. Asian Pac J Cancer Prev. 2013; 14:7085–7090.

11. Lind EF, Ohashi PS. Mir-155, a central modulator of T-cell responses. Eur J Immunol. 2014; 44:11–15.

12. Zheng Y, Xiong S, Jiang P, Liu R, Liu X, Qian J, et al. Glucocorticoids inhibit lipopolysaccharide-mediated inflammatory response by downregulating microRNA-155: a novel anti-inflammation mechanism. Free Radic Biol Med. 2012; 52:1307–1317.

13. Bera A, VenkataSubbaRao K, Manoharan MS, Hill P, Freeman JW. A miRNA signature of chemoresistant mesenchymal phenotype identifies novel molecular targets associated with advanced pancreatic cancer. PLoS One. 2014; 9:e106343.

14. Chen L, Jiang K, Jiang H, Wei P. miR-155 mediates drug resistance in osteosarcoma cells via inducing autophagy. Exp Ther Med. 2014; 8:527–532.

15. Pareek S, Roy S, Kumari B, Jain P, Banerjee A, Vrati S. MiR-155 induction in microglial cells suppresses Japanese encephalitis virus replication and negatively modulates innate immune responses. J Neuroinflammation. 2014; 11:97.

16. Zhu J, Chen T, Yang L, Li Z, Wong MM, Zheng X, et al. Regulation of microRNA-155 in atherosclerotic inflammatory responses by targeting MAP3K10. PLoS One. 2012; 7:e46551.

17. Anzi S, Finkin S, Shaulian E. Transcriptional repression of c-Jun's E3 ubiquitin ligases contributes to c-Jun induction by UV. Cell Signal. 2008; 20:862–871.

18. Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014; 15:429.
19. Ayaz L, Görür A, Yaroğlu HY, Ozcan C, Tamer L. Differential expression of microRNAs in plasma of patients with laryngeal squamous cell carcinoma: potential early-detection markers for laryngeal squamous cell carcinoma. J Cancer Res Clin Oncol. 2013; 139:1499–1506.

20. Sun X, Song Y, Tai X, Liu B, Ji W. MicroRNA expression and its detection in human supraglottic laryngeal squamous cell carcinoma. Biomed Rep. 2013; 1:743–746.

21. Wu TY, Zhang TH, Qu LM, Feng JP, Tian LL, Zhang BH, et al. MiR-19a is correlated with prognosis and apoptosis of laryngeal squamous cell carcinoma by regulating TIMP-2 expression. Int J Clin Exp Pathol. 2013; 7:56–63.
22. Jones K, Nourse JP, Keane C, Bhatnagar A, Gandhi MK. Plasma microRNA are disease response biomarkers in classical Hodgkin lymphoma. Clin Cancer Res. 2014; 20:253–264.

23. Vimalraj S, Miranda PJ, Ramyakrishna B, Selvamurugan N. Regulation of breast cancer and bone metastasis by microRNAs. Dis Markers. 2013; 35:369–387.

24. Gao F, Chang J, Wang H, Zhang G. Potential diagnostic value of miR-155 in serum from lung adenocarcinoma patients. Oncol Rep. 2014; 31:351–357.

25. Hui AB, Lenarduzzi M, Krushel T, Waldron L, Pintilie M, Shi W, et al. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res. 2010; 16:1129–1139.

26. Gasparini P, Cascione L, Fassan M, Lovat F, Guler G, Balci S, et al. microRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget. 2014; 5:1174–1184.

27. Kopp KL, Ralfkiaer U, Nielsen BS, Gniadecki R, Woetmann A, Ødum N, et al. Expression of miR-155 and miR-126 in situ in cutaneous T-cell lymphoma. APMIS. 2013; 121:1020–1024.

28. Sochor M, Basova P, Pesta M, Dusilkova N, Bartos J, Burda P, et al. Oncogenic microRNAs: miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. BMC Cancer. 2014; 14:448.

29. Sun J, Shi H, Lai N, Liao K, Zhang S, Lu X. Overexpression of microRNA-155 predicts poor prognosis in glioma patients. Med Oncol. 2014; 31:911.

30. Zhao XD, Zhang W, Liang HJ, Ji WY. Overexpression of miR-155 promotes proliferation and invasion of human laryngeal squamous cell carcinoma via targeting SOCS1 and STAT3. PLoS One. 2013; 8:e56395.
31. Xu L, Li M, Wang M, Yan D, Feng G, An G. The expression of microRNA-375 in plasma and tissue is matched in human colorectal cancer. BMC Cancer. 2014; 14:714.

32. Zhang R, Wang W, Li F, Zhang H, Liu J. MicroRNA-106b~25 expressions in tumor tissues and plasma of patients with gastric cancers. Med Oncol. 2014; 31:243.

33. Herberth G, Bauer M, Gasch M, Hinz D, Röder S, Olek S, et al. Maternal and cord blood miR-223 expression associates with prenatal tobacco smoke exposure and low regulatory T-cell numbers. J Allergy Clin Immunol. 2014; 133:543–550.

34. Basso K, Schneider C, Shen Q, Holmes AB, Setty M, Leslie C, et al. BCL6 positively regulates AID and germinal center gene expression via repression of miR-155. J Exp Med. 2012; 209:2455–2465.

35. Sandhu SK, Volinia S, Costinean S, Galasso M, Neinast R, Santhanam R, et al. miR-155 targets histone deacetylase 4 (HDAC4) and impairs transcriptional activity of B-cell lymphoma 6 (BCL6) in the Eµ-miR-155 transgenic mouse model. Proc Natl Acad Sci U S A. 2012; 109:20047–20052.

36. Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005; 102:3627–3632.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download