Abstract
The trends and types of carbapenemase-producing Gram-negative bacilli were analyzed from clinical specimens collected between 2005 and 2012 at a Korean teaching hospital. The proportions of carbapenem-resistant Acinetobacter spp. increased markedly to 66%. Metallo-β-lactamase producers significantly decreased and the majority shifted from the blaVIM-2 type to the blaIMP-1 type.
Carbapenems such as imipenem and meropenem are first-line drugs in the treatment of serious infections caused by multidrug-resistant Pseudomonas aeruginosa, Acinetobacter spp., and Enterobacteriaceae. However, in recent years, imipenem resistance in P. aeruginosa and Acinetobacter spp. has been increasing steadily around the world, and in Korea, these resistance rates reached 22% and 64%, respectively, in 2011.1 Among several mechanisms of carbapenem resistance, acquired metallo-β-lactamases (MBLs) have a more serious impact as the enzymes confer a high level of resistance and the genes can be transferred horizontally.2,3 Among acquired MBLs, VIM-type and IMP-type enzymes are the most common types of MBLs with worldwide distribution,4 and the VIM-2 type has been highly prevalent in Korea.5,6 In our previous study in 2003-2004, imipenem-nonsusceptible P. aeruginosa isolates carrying the blaVIM-2 allele were highly prevalent, and the incidence of Acinetobacter spp. carrying the blaVIM-2 allele had increased compared to those carrying the blaIMP-1 allele.5 The aim of the present study was to determine the trends of carbapenem-resistant and MBL-producing Gram-negative bacilli over the past 8 years in a Korean teaching hospital with more than 2000 beds. This is a unique report on the long term trend of carbapenem-resistant and MBL-producing Gram-negative bacilli isolated in a single hospital.
In total, non-duplicated clinical isolates of 12650 P. aeruginosa, 1096 other Pseudomonas spp., and 7650 Acinetobacter spp. in addition to 14026 Klebsiella pneumoniae, 6110 Enterobacter cloacae, and 3162 Serratia marcescens isolates among Enterobacteriaceae were recovered from patients at the hospital from 2005 to 2012 (Table 1). The isolates were identified by conventional methods using ATB 32 GN or VITEK-2 systems (bioMerieux, Marcy-l'E'toile, France). Antimicrobial susceptibilities were determined using the disk-diffusion method or the VITEK-2 system (bioMerieux). The Clinical and Laboratory Standards Institute 2010 breakpoints were used after January 2011.7 The modified Hodge test (MHT) and the imipenem and ethylenediaminetetraacetic acid sodium mercaptoacetic acid double disk potentiation (IEDDP) test were conducted to screen for MBL producers in imipenem- or meropenem-nonsusceptible isolates.8 Polymerase chain reaction (PCR) was performed to detect and sequence blaVIM-2-like, blaIMP-1-like and blaSIM-1-like genes.9,10 In carbapenem-nonsusceptible strains showing MHT-positive yet the aforementioned PCR negative results, blaNDM and blaKPC genes were also screened by PCR.11,12 The nucleotide sequences of PCR-generated amplicons were analyzed for the representative strains. XbaI- or SmaI-digested genomic DNAs from 75 P. aeruginosa, 8 P. putida, and 109 Acinetobacter spp., all of which were randomly selected MBL-producing spp. isolated in 2005-2006, were separated by pulsed field gel electrophoresis (PFGE) using a CHEF-DR II system (Bio-Rad, Hercules, CA, USA) and BioNumerics software v. 5.10 (Applied Maths, Sint-Martens-Latem, Belgium). PFGE banding pattern clustering with an 80% similarity threshold was determined using the Dice coefficients and the unweighted pair group method using arithmetic averages using Molecular Analyst Fingerprinting Software (Bio-Rad). Related clones with one or two independent genetic events were designated as subtype numbers in Table 2. The S1-digested DNA of randomly selected P. aeruginosa, Pseudomonas putida, and Acinetobacter spp. strains carrying the blaVIM-2 allele were blotted onto nylon membranes (Bio-Rad) and hybridized with blaVIM-2 gene probes to observe the differences in the plasmids carrying the gene.
While annual imipenem resistance rates in P. aeruginosa increased from 14% in 2003 to 29% in 2009, they decreased from 58% in 2003 to 33% in 2012 in other Pseudomonas spp.; in Acinetobacter spp., resistance rates increased steeply from 13% in 2003 to 66% in 2012.5 However, the imipenem resistance rates in K. pneumoniae, E. cloacae, and S. marcescens isolates remained at less than 1%. The positive rates of MHT were highly variable depending on the species, being only 13% in P. aeruginosa isolates, yet reaching 69% in other Pseudomonas spp. and 94% in Acinetobacter spp. These findings suggest that the majority of other Pseudomonas and Acinetobacter spp. isolates clearly produced the carbapenem-hydrolyzing enzymes to gain their resistance to carbapenem, while P. aeruginosa isolates obtained other resistance mechanisms such as AmpC or extended spectrum β-lactamase and porin loss, as previously reported.13,14
Among the carbapenem-nonsusceptible isolates, MBL producers included only 6% (243 of 4077) of P. aeruginosa spp. and only 7% (262 of 3998) of Acinetobacter spp. Interestingly, 157 of 247 (64%) of the other Pseudomonas spp. produced MBL, and almost all of them (152 of 157) were P. putida, which was a much higher incidence than those of the other species. These findings suggest that P. putida can be a reservoir for MBL, as previously described.15 Acinetobacter spp. with different carbapenem-resistance mechanisms, such as OXA-type β-lactamases, have become prevalent.16
In P. aeruginosa, the incidence of MBL producers increased until the mid-2000s in this study, as shown in Japan,17 while in more recent years, these isolates gradually decreased in this study, as described in a previous report.18 Likewise, Cavalcanti, et al.19 reported that a higher prevalence of MBL-producing P. aeruginosa was observed in 2002-2003 in Brazil, while the level decreased significantly in 2008-2009, suggesting that the resistance to carbapenems by these recent P. aeruginosa isolates was not due to the spread of MBL-positive clones. In this study, P. aeruginosa isolates carrying a blaVIM-2-like gene were highly prevalent, comprising 90% to 100% of the P. aeruginosa strains in 2003 to 2004,5 although they were reduced to 34% while those with blaIMP-1-like genes increased to 69% in 2010.
Among MBL-producing Acinetobacter spp. isolates, the prevalence of blaIMP-1-like genes also increased to 85% in 2012. The range of strains carrying blaSIM-1-like genes remained low. Two isolates carrying blaNDM-1-like genes that were isolated in 2011 were identified as A. pitti and A. guillouiae. To our knowledge, this is the first report of a clinical isolate of A. guillouiae carrying the blaNDM-1-like gene. Most of the other Pseudomonas spp. isolates carrying MBL genes were identified as P. putida, and their MBL genes were blaVIM-2-like.
Among the 254 carbapenem-nonsusceptible Enterobacteriaceae, only five isolates, two K. pneumoniae, two E. cloacae, and one S. marcescens, produced MBL, suggesting that the major carbapenem resistance mechanism in Enterobacteriaceae was not MBL. Our results support previous reports that suggested that carbapenem resistance in Enterobacteriaceae was comediated with AmpC beta-lactamase and outer membrane protein loss in K. pneumoniae, E. cloacae, and S. marcescens.20,21,22
PFGE analysis revealed that the pulsotypes of IMP-6- and VIM-2-producing P. aeruginosa strains were clearly separated. The major pulsotypes in the IMP-6-producing P. aeruginosa were A2 and A3, while those in the VIM-2-producing P. aeruginosa were A1 and C1 types (Table 2). Likewise, the pulsotypes of Acinetobacter spp. isolates obviously differed according to MBL type (Table 2). These findings suggest that the plasmids carrying the MBL gene are not promiscuous, although they do have clone preference. Interestingly, Acinetobacter spp. isolates in the E1, H1, I4, J3, N1, and Q subgroups showed identical PFGE patterns, despite the differences in species. This suggests that the identification of Acinetobacter species is important for evaluating clonal outbreaks in hospital settings, as the misinterpretation of a clonal outbreak occurred among the different species.
Other mechanisms may block the cross-over of resistance plasmids between clones. Further study is warranted to elucidate this supposition. The hybridization of S1-digested DNA showed that the sizes of blaVIM-2 gene-carrying plasmids in P. aeruginosa, P. putida, and Acinetobacter spp. isolates were diverse (Supplementary Fig. 1, only online). It is noteworthy that the blaVIM-2 gene-carrying plasmids in Acinetobacter spp. were in multimer forms, indicating that the plasmids did not replicate themselves in the same way as with P. aeruginosa. Further plasmid sequence analysis using massive parallel sequencing technology has been undertaken.
In conclusion, MBL-producing clinical isolates of P. aeruginosa and Acinetobacter spp. were reduced, and carbapenemase-producing Enterobacteriaceae were found to be rare in Korea. Continuous surveillance studies and further deep sequencing are necessary to understand the dissemination mechanism of the carbapenem-nonsusceptible Gram-negative bacilli isolates in order to control their spread.
Figures and Tables
Table 1
Annual Imipenem Resistance Rates and MBL-Producing Clinical Isolates over 8 Years
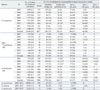
Table 2
Pulsotypes of 109 MBL-Producing Acinetobacter spp. and 75 P. aeruginosa Isolates

ACKNOWLEDGEMENTS
This study was supported by faculty research grant 6-2014-0039 from Yonsei University College of Medicine for 2014.
References
1. Yong D, Shin HB, Kim YK, Cho J, Lee WG, Ha GY, et al. Increase in the prevalence of carbapenem-resistant Acinetobacter isolates and ampicillin-resistant non-typhoidal Salmonella species in Korea: a KONSAR study conducted in 2011. Infect Chemother. 2014; 46:84–93.

2. Walsh TR. Clinically significant carbapenemases: an update. Curr Opin Infect Dis. 2008; 21:367–371.

3. Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect. 2006; 12:826–836.

4. Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-beta-lactamases: the quiet before the storm? Clin Microbiol Rev. 2005; 18:306–325.
5. Yong D, Choi YS, Roh KH, Kim CK, Park YH, Yum JH, et al. Increasing prevalence and diversity of metallo-beta-lactamases in Pseudomonas spp., Acinetobacter spp., and Enterobacteriaceae from Korea. Antimicrob Agents Chemother. 2006; 50:1884–1886.

6. Lim Y, Lee Y, Seo Y, Yum JH, Yong D, Lee K, et al. Loss of blaVIM-2 and blaIMP-1 during the storage of gram-negative bacilli, antimicrobial susceptibility of the gene-lost strain, and location of the gene in the cell. Ann Clin Microbiol. 2013; 16:120–125.

7. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing: twentieth informational supplement. M100-S20-U. Wayne, PA: CLSI;2010.
8. Lee K, Lim YS, Yong D, Yum JH, Chong Y. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-beta-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2003; 41:4623–4629.

9. Lee K, Lee WG, Uh Y, Ha GY, Cho J, Chong Y, et al. VIM- and IMP-type metallo-beta-lactamase-producing Pseudomonas spp. and Acinetobacter spp. in Korean hospitals. Emerg Infect Dis. 2003; 9:868–871.

10. Lee K, Yum JH, Yong D, Lee HM, Kim HD, Docquier JD, et al. Novel acquired metallo-beta-lactamase gene, blaSIM-1, in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob Agents Chemother. 2005; 49:4485–4491.

11. Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009; 53:5046–5054.

12. Smith Moland E, Hanson ND, Herrera VL, Black JA, Lockhart TJ, Hossain A, et al. Plasmid-mediated, carbapenem-hydrolysing beta-lactamase, KPC-2, in Klebsiella pneumoniae isolates. J Antimicrob Chemother. 2003; 51:711–714.

13. Rodríguez-Martínez JM, Poirel L, Nordmann P. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009; 53:4783–4788.

14. Quale J, Bratu S, Gupta J, Landman D. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother. 2006; 50:1633–1641.

15. Juan C, Zamorano L, Mena A, Albertí S, Pérez JL, Oliver A. Metallo-beta-lactamase-producing Pseudomonas putida as a reservoir of multidrug resistance elements that can be transferred to successful Pseudomonas aeruginosa clones. J Antimicrob Chemother. 2010; 65:474–478.

17. Kouda S, Ohara M, Onodera M, Fujiue Y, Sasaki M, Kohara T, et al. Increased prevalence and clonal dissemination of multidrug-resistant Pseudomonas aeruginosa with the blaIMP-1 gene cassette in Hiroshima. J Antimicrob Chemother. 2009; 64:46–51.

18. Lee JY, Ko KS. OprD mutations and inactivation, expression of efflux pumps and AmpC, and metallo-β-lactamases in carbapenem-resistant Pseudomonas aeruginosa isolates from South Korea. Int J Antimicrob Agents. 2012; 40:168–172.

19. Cavalcanti FL, Almeida AC, Vilela MA, Morais MM, Morais Junior MA. Changing the epidemiology of carbapenem-resistant Pseudomonas aeruginosa in a Brazilian teaching hospital: the replacement of São Paulo metallo-β-lactamase-producing isolates. Mem Inst Oswaldo Cruz. 2012; 107:420–423.

20. Suh B, Bae IK, Kim J, Jeong SH, Yong D, Lee K. Outbreak of meropenem-resistant Serratia marcescens comediated by chromosomal AmpC beta-lactamase overproduction and outer membrane protein loss. Antimicrob Agents Chemother. 2010; 54:5057–5061.

21. Shin SY, Bae IK, Kim J, Jeong SH, Yong D, Kim JM, et al. Resistance to carbapenems in sequence type 11 Klebsiella pneumoniae is related to DHA-1 and loss of OmpK35 and/or OmpK36. J Med Microbiol. 2012; 61(Pt 2):239–245.

22. Lee Y, Choi H, Yum JH, Kang G, Bae IK, Jeong SH, et al. Molecular mechanisms of carbapenem resistance in Enterobacter cloacae clinical isolates from Korea and clinical outcome. Ann Clin Lab Sci. 2012; 42:281–286.
Supplementary Material
Supplementary Fig. 1.
(A) Pulsed field gel electrophoresis of whole genomic DNA of VIM-2-producing P. aeruginosa (lanes 2 to 16), P. putida (lanes 17 to 22), and Acinetobacter spp. isolates (lanes 23 to 27) digested with S1 nuclease. (B) Southern blot hybridization with blaVIM-2 gene probe. Lane M, lambda ladder (Bio-Rad) as a marker (kb). The genomic DNA of P. aeruginosa stains with high endogenous DNase activities were degraded not to show positive bands (lanes 2, 3, 4, 6, 7, 8, 9, 11, 12, 13, and 15).




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download